Case Studies in Library Design¶
Info
This chapter provides specifics on focused and diverse libraries. The examples chart the different steps involved in library design including the definition of the chemical reaction, the selection of building blocks, library optimization and the reduction of the virtual library to a manageable set of compounds. The results are presented and discussed.
Number of Pages: 53 (±1 hours read)
Last Modified: September 2008
Prerequisites: None
Case Study-1 : CDK2 Inhibitors¶
Purine Scaffold as a Source of Bioactive Molecules¶
A great number of proteins, such as the kinases, DNA polymerases, ATPases, GTPases, purine receptors etc..., depend on purine-containing ligands for their function. Guanine and adenine are examples of purine-based cofactors involved in many biological systems. For this reason, the Schultz-Meijer groups suggested that purine-based libraries may be of high informational content and useful as a source of probes for analyzing complex cellular processes and generating leads for drug development. A library design strategy is presented here that led to the discovery of potent ligands such as purvanalol A, B and amino-purvanalol which are active at the nano-molar level.
CDK2 Biological Target and Known Inhibitors¶
In order to test their hypothesis, purine libraries were developed to address the inhibition of cyclin-dependent kinases (CDKs) that are important in regulating the cell cycle. Compounds such as olomoucine and roscovitine were already known, but their CDK inhibitory activities were in the micromolar range only.
Diverse 2,6,9-trisubstituted Purine Libraries¶
A solid-phase synthesis was conceived as outlined below, which permitted the introduction of substituents at the 2, 6 and 9 positions of the purine scaffold by immobilizing it through the amino group in position 6.
Substituent Design¶
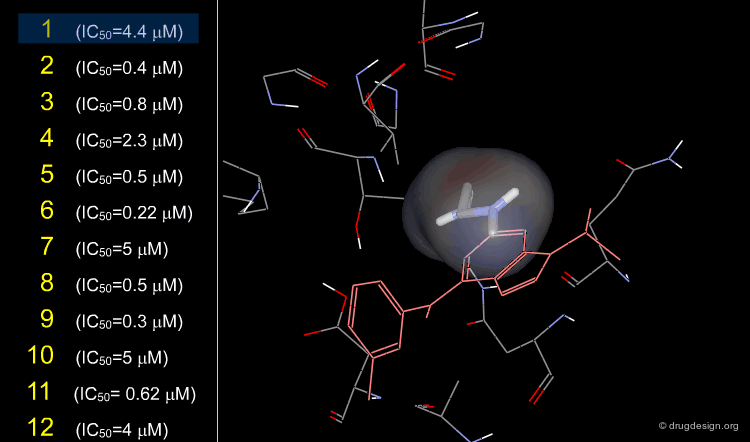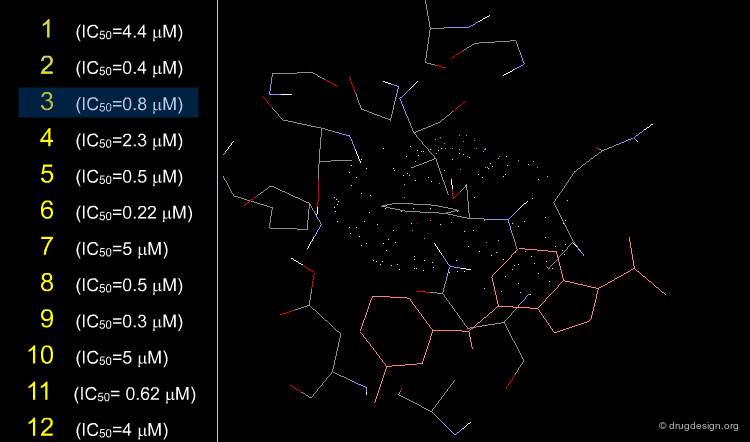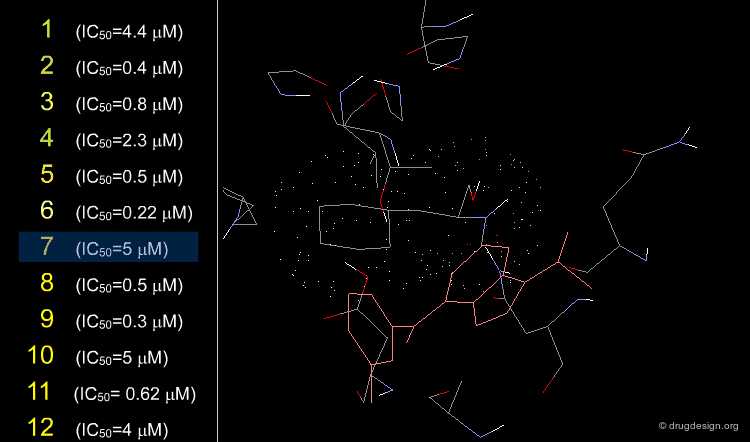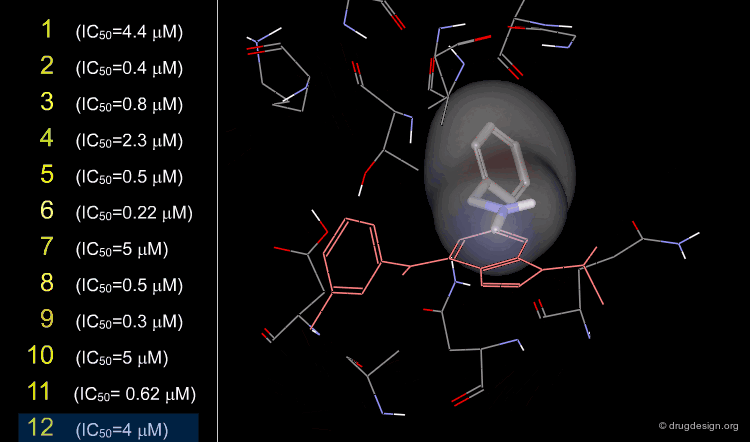
The X-ray structures of ligands bound to CDK2 permitted the design of substituents at the 2, 6 and 9 positions; they were selected based on their steric and electronic interactions with the protein. Moreover the binding modes of known inhibitors provided useful information about functional groups that could be accommodated in the catalytic site.
Additivity of the Biological Effects¶
The process relied on the additivity of the substituents, an assumption that proved to be valid. It made it possible to reduce the number of compounds; for example, in the evaluation of 20 substituents at each site it requires only three sets of 20 compounds rather than one set of 8000.
Browser of Substituents at the C-2 Position¶
Example of substituents that were considered at the C2-position are available in this browser (click the molecules and try to interpret the SAR). This illustrates the power of integrating combinatorial chemistry and structure based drug design.
Browser of Substituents at the C-6 Position¶
Example of substituents that were considered at the C6 position are available in this browser (click the molecules and try to interpret the SAR).
Successive Rounds¶
The synthetic approach was iteratively applied to the 2, 6 and 9 positions with biological testing in parallel. Several rounds were conducted by fixing some positions with a given group and varying the others for optimization. It started by the exploration at the N9 position that showed that only a small alkyl group could be accommodated, the best activities were achieved with an isopropyl substituent. The optimization at the C2 and C6 positions was further developed with an isopropyl at N9.
Library Results¶
An informative purine library constructed in conjunction with a structure-based approach allowed for the discovery of potent lead compounds such as purvanalol A, B and aminopurvanalol, active at the nanomolar level. The molecules proved to arrest cellular proliferation and were further exploited to analyze the mechanisms associated to the cellular process.
Case Study-2 : DHFR Inhibitors¶
Diaminopyrimidines DHFR Inhibitors¶
The dihydrofolate reductase enzyme (DHFR) has been clinically proven to be a relevant target for chemotherapy and the trimethoprim (TMP) inhibitor is often used as a therapeutic agent against Gram-negative pathogens; however, there is a need to find antibacterial agents against the highly resistant Gram-positive organisms S. aureus and S. pneumoniae. The Roche group identified highly active compounds such as RO-64-5781; but they could not be developed further due to their high protein binding and low solubility, which were probably caused by their high molecular weight and lipophilicity. In the following pages we present a library design strategy that have led to the discovery of novel and soluble structures with great potential.
Author
Martin Stahl Pharmaceutical Division, F. Hoffmann-La-Roche Ltd. CH-4070, Basle, Switzerland
articles
Novel dihydrofolate reductase inhibitors. Structure-based vs. diversity-based library design and high-throughput synthesis and screening Wyss, P. C., P. R. Gerber, et al. J Med Chem 46 2003
Soluble Diaminopyrimidine Scaffold¶
The RO-64-5781 and TMP molecules have a 2,4-diaminopyrimidine scaffold with a lipophilic benzyl group at the 5-position. The Roche group hypothesized that 2,4-diaminopyrimidine derivatives incorporating basic N-disubstituted aminomethyl small residues at the 5-position would be less lipophilic than RO-64-5781 and might lead to a second generation of inhibitors having better solubility properties.
Author
Martin Stahl Pharmaceutical Division, F. Hoffmann-La-Roche Ltd. CH-4070, Basle, Switzerland
articles
Novel dihydrofolate reductase inhibitors. Structure-based vs. diversity-based library design and high-throughput synthesis and screening Wyss, P. C., P. R. Gerber, et al. J Med Chem 46 2003
Design of 2,4-Diaminopyrimidine Library¶
A parallel synthetic scheme was devised, based on the availability of 9448 secondary amines of the company's proprietary library. This is a clean reaction sequence that didn't even require compound purification.
Author
Martin Stahl Pharmaceutical Division, F. Hoffmann-La-Roche Ltd. CH-4070, Basle, Switzerland
articles
Novel dihydrofolate reductase inhibitors. Structure-based vs. diversity-based library design and high-throughput synthesis and screening Wyss, P. C., P. R. Gerber, et al. J Med Chem 46 2003
Structure-Based Design Strategy¶
To guide the synthetic program, a structure-based strategy was adopted that aimed at docking and scoring all the molecules of the library into the DHFR targets. Molecules having the best scores are selected for chemical synthesis and biological tests.
Author
Martin Stahl Pharmaceutical Division, F. Hoffmann-La-Roche Ltd. CH-4070, Basle, Switzerland
articles
Novel dihydrofolate reductase inhibitors. Structure-based vs. diversity-based library design and high-throughput synthesis and screening Wyss, P. C., P. R. Gerber, et al. J Med Chem 46 2003
3D Structural Data Available¶
About 30 complexes between S. aureus DHFR and first generation inhibitors were solved at Roche and other complexes were also available in the PDB. They showed no significant structural variations in the catalytic site of the enzyme. A homology model constructed for S. pneumoniae DHFR revealed a binding site similar to that of S. aureus DHFR. The use of a single crystal structure of S. aureus DHFR was therefore assumed to be sufficient for a structure-based selection of compounds against the two targets.
Author
Martin Stahl Pharmaceutical Division, F. Hoffmann-La-Roche Ltd. CH-4070, Basle, Switzerland
articles
Novel dihydrofolate reductase inhibitors. Structure-based vs. diversity-based library design and high-throughput synthesis and screening Wyss, P. C., P. R. Gerber, et al. J Med Chem 46 2003
2,4-Diaminopyrimidine Anchorage to DHFR¶
Analysis of the complexes of 2,4-diaminopyrimidine derivatives of the first generation with S. aureus revealed a common anchorage with an extended hydrogen bond network between the 2,4-diaminopyrimidine and the amino acids Phe-92, Leu-5 and Asp-27. All the molecules in the virtual library should be docked with the constraint that the 2,4-diaminopyrimidine fragment must have a fixed position.
Author
Martin Stahl Pharmaceutical Division, F. Hoffmann-La-Roche Ltd. CH-4070, Basle, Switzerland
articles
Novel dihydrofolate reductase inhibitors. Structure-based vs. diversity-based library design and high-throughput synthesis and screening Wyss, P. C., P. R. Gerber, et al. J Med Chem 46 2003
Docking of the Virtual Library¶
Of the 9448 Roche proprietary library of secondary amines, 3909 compounds were excluded for pharmacokinetic reasons (solubility, pKa, clogP and molecular weight) leading to a virtual library of 4439 diaminopyrimidines that could be assessed by docking. The molecules were generated with an in-house library enumeration program and single 3D conformations were constructed with the Corina program. The docking program FlexX was used with an improved scoring function developed at Roche for virtual screening applications.
Author
Martin Stahl Pharmaceutical Division, F. Hoffmann-La-Roche Ltd. CH-4070, Basle, Switzerland
articles
Novel dihydrofolate reductase inhibitors. Structure-based vs. diversity-based library design and high-throughput synthesis and screening Wyss, P. C., P. R. Gerber, et al. J Med Chem 46 2003
CORINA: Automatic generation of 3D atomic coordinates for organic molecules Gasteiger J, Rudolph C, Sadowski J. Tetrahedron Computer Methodology 3 1990
Modification of the scoring function in FlexX for virtual screening applications Stahl M Perspect. Drug Discovery Des 20 2000
Selection and Synthesis¶
A library of 252 molecules with top-ranking docking scores was defined and the molecules were all synthesized. Some of the structures found in this library are displayed here; they contain bi-, tri and tetracyclic amine structures.
Author
Martin Stahl Pharmaceutical Division, F. Hoffmann-La-Roche Ltd. CH-4070, Basle, Switzerland
articles
Novel dihydrofolate reductase inhibitors. Structure-based vs. diversity-based library design and high-throughput synthesis and screening Wyss, P. C., P. R. Gerber, et al. J Med Chem 46 2003
Biological Tests¶
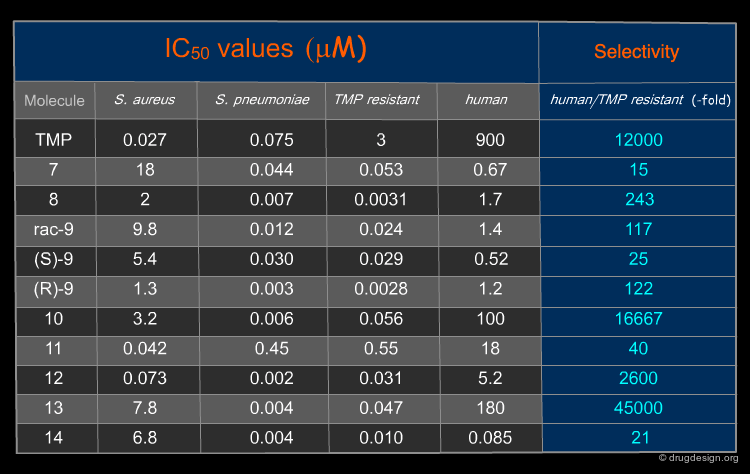
The 252 molecules in the library were tested against isolated DHFR from S. aureus, S. pneumoniae and TMP resistant DHFR from streptococcus pneumoniae. 54 hits were found, which corresponded to a success rate of 21%. The IC50s and the selectivity of some hits are shown here and compared with the values found for trimethoprim (TMP).
Author
Martin Stahl Pharmaceutical Division, F. Hoffmann-La-Roche Ltd. CH-4070, Basle, Switzerland
articles
Novel dihydrofolate reductase inhibitors. Structure-based vs. diversity-based library design and high-throughput synthesis and screening Wyss, P. C., P. R. Gerber, et al. J Med Chem 46 2003
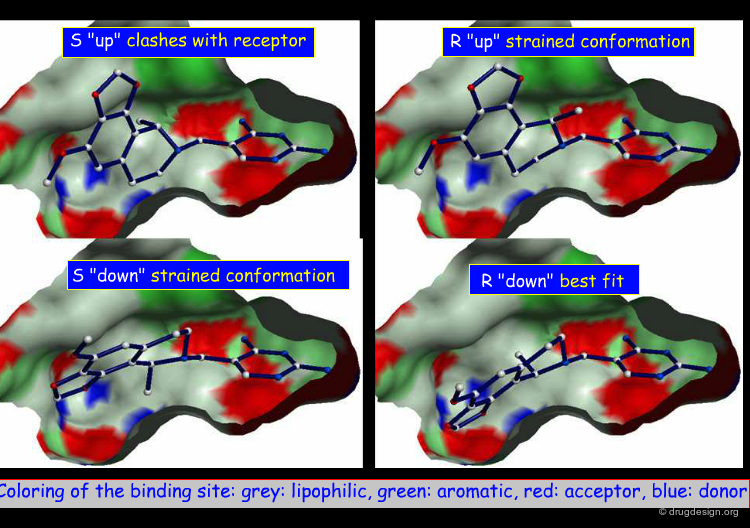
Detailed Analysis of Binding Mode¶
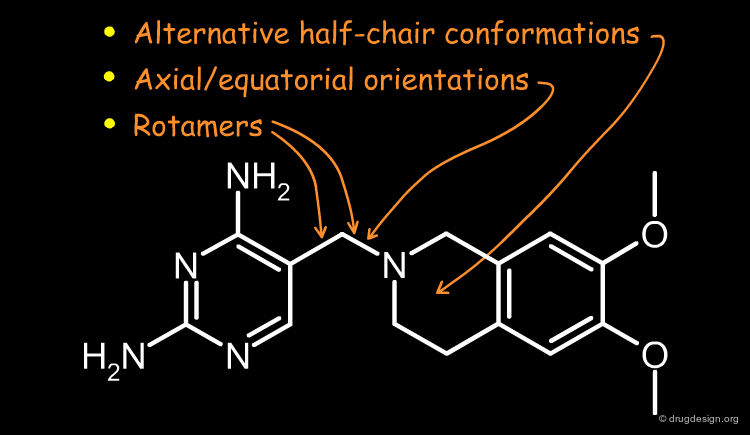
A more precise analysis of the binding mode of the hits was required to help achieve their optimization. The 2,4-diaminopyrimidine tetrahydroisoquinoline fragment was common to many of the hits obtained, and its full conformational analysis was made. The analyses revealed that only half-chair equatorial conformations could be accommodated in the pocket, leading to two alternatives for the binding mode: the tetrahydroisoquinoline could either be "up" or "down".
Author
Martin Stahl Pharmaceutical Division, F. Hoffmann-La-Roche Ltd. CH-4070, Basle, Switzerland
articles
Novel dihydrofolate reductase inhibitors. Structure-based vs. diversity-based library design and high-throughput synthesis and screening Wyss, P. C., P. R. Gerber, et al. J Med Chem 46 2003
Enantiomers with Different Activities¶
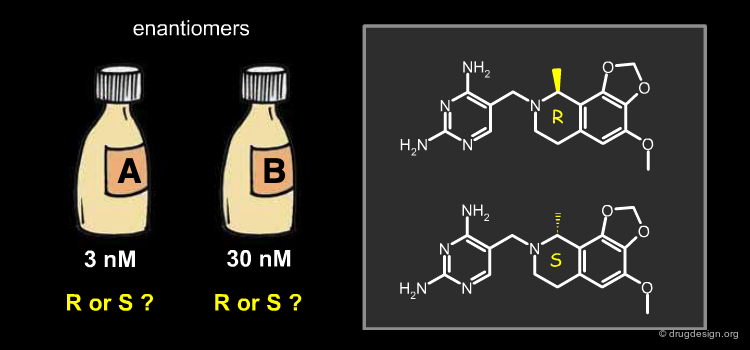
To determine the exact binding mode (up or down) of the molecules, the authors analyzed compound 9 that had a stereo center with one enantiomer that was more active than the other. This study showed that only the R configuration with the "down" orientation could fit optimally to the receptor. This is explained on the next page.
Author
Martin Stahl Pharmaceutical Division, F. Hoffmann-La-Roche Ltd. CH-4070, Basle, Switzerland
articles
Novel dihydrofolate reductase inhibitors. Structure-based vs. diversity-based library design and high-throughput synthesis and screening Wyss, P. C., P. R. Gerber, et al. J Med Chem 46 2003
Binding Mode and Absolute Stereochemistry¶
The docking analyses showed that the R "down" binding mode fit optimally in the binding pocket with the axial methyl group accessing a hitherto unexplored sub-pocket. The S "down" and R "up" possibilities were discarded because of their high energy, and the S "up" possibility was eliminated due to clashes with the receptor. Subsequent X-ray experiments confirmed the modeling predictions.
Author
Martin Stahl Pharmaceutical Division, F. Hoffmann-La-Roche Ltd. CH-4070, Basle, Switzerland
articles
Novel dihydrofolate reductase inhibitors. Structure-based vs. diversity-based library design and high-throughput synthesis and screening Wyss, P. C., P. R. Gerber, et al. J Med Chem 46 2003
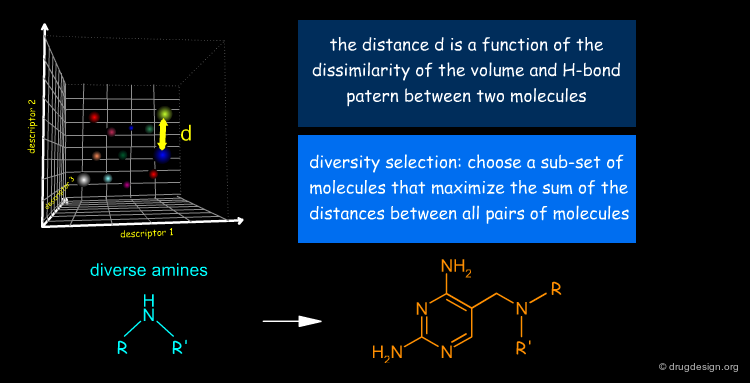
Diversity-Based Strategy¶
Alongside the structure-based selection of candidate inhibitors, the Roche team pursued a diversity-based strategy with the aim of comparing the success rates obtained with the two approaches. Assessment of diversity was based on the volume occupied by the molecules and the distribution of hydrogen-bond capabilities.
Author
Martin Stahl Pharmaceutical Division, F. Hoffmann-La-Roche Ltd. CH-4070, Basle, Switzerland
articles
Novel dihydrofolate reductase inhibitors. Structure-based vs. diversity-based library design and high-throughput synthesis and screening Wyss, P. C., P. R. Gerber, et al. J Med Chem 46 2003
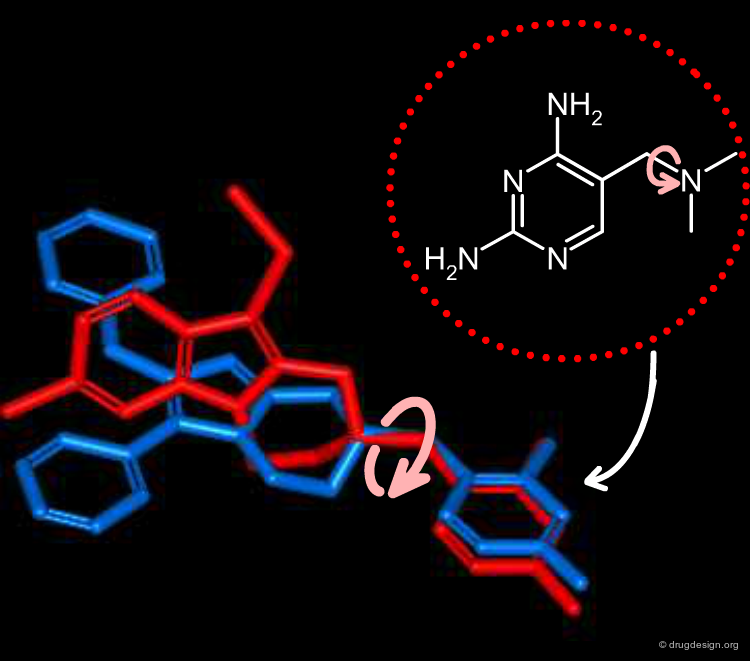
Selection Based on Diversity of Pair Overlaps¶
To classify the molecules according to their volume, all pairs of molecules were aligned by keeping the 2,4-diaminopyrimidine fragments superimposed as indicated below. The amine substituents were rotated around the newly formed C-N bond to maximize the overlap. The volume overlap and H-bond properties led to pairwise similarity scores (Moloc program).
Author
Martin Stahl Pharmaceutical Division, F. Hoffmann-La-Roche Ltd. CH-4070, Basle, Switzerland
articles
Novel dihydrofolate reductase inhibitors. Structure-based vs. diversity-based library design and high-throughput synthesis and screening Wyss, P. C., P. R. Gerber, et al. J Med Chem 46 2003
Selection of Molecules and Biological Tests¶
All the pairs or molecules were organized by hierarchical clustering and 501 diverse representative compounds were selected to represent the chemical space spanned by the library. They were all synthesized and tested. Only 17 molecules were found with some inhibitory activities, which corresponds to a success rate of only 3%. No potent inhibitors resulted from this approach.
Author
Martin Stahl Pharmaceutical Division, F. Hoffmann-La-Roche Ltd. CH-4070, Basle, Switzerland
articles
Novel dihydrofolate reductase inhibitors. Structure-based vs. diversity-based library design and high-throughput synthesis and screening Wyss, P. C., P. R. Gerber, et al. J Med Chem 46 2003
Structure-Based vs. Diversity-Based Strategy¶
This work demonstrates the vast superiority of structure-based selection as compared to selection by diversity. More highly active compounds were obtained through docking and all the interesting new compound classes were covered in the docking libraries.
Author
Martin Stahl Pharmaceutical Division, F. Hoffmann-La-Roche Ltd. CH-4070, Basle, Switzerland
articles
Novel dihydrofolate reductase inhibitors. Structure-based vs. diversity-based library design and high-throughput synthesis and screening Wyss, P. C., P. R. Gerber, et al. J Med Chem 46 2003
Efficiency of the Structure-Based Selection¶
A control experiment was conducted to test the efficiency of the structure-based selection: 150 structures that could not be docked as well as another 150 structures with low docking score were selected and synthesized. Out of these, only one weakly active compound was identified. Since the team had a clean reaction that didn't require compound purification, they could afford to make and test all the compounds they wanted to test. This is one of the rare published study where a library of compounds predicted to be inactive was actually made and tested.
Author
Martin Stahl Pharmaceutical Division, F. Hoffmann-La-Roche Ltd. CH-4070, Basle, Switzerland
articles
Novel dihydrofolate reductase inhibitors. Structure-based vs. diversity-based library design and high-throughput synthesis and screening Wyss, P. C., P. R. Gerber, et al. J Med Chem 46 2003
Summary¶
The project was aimed at discovering antibacterial agents to fight the highly resistant Gram-positive organisms S. aureus and S. pneumoniae. Libraries were compiled using a structure-based and a diversity-based strategy for compound selection. Two strategies were considered because at the time neither the team members nor many of their coworkers had recognized the true advantages of structure-based virtual screening. This study led to the identification of a new generation of potent and soluble inhibitors, but unfortunately the project was halted due to the closure of all of anti-infectives research at Roche.
Author
Martin Stahl Pharmaceutical Division, F. Hoffmann-La-Roche Ltd. CH-4070, Basle, Switzerland
What can we Learn from this Study ?¶
For the Roche team, the key lessons learned over the course of this project are listed below.
Author
Martin Stahl Pharmaceutical Division, F. Hoffmann-La-Roche Ltd. CH-4070, Basle, Switzerland
articles
Novel dihydrofolate reductase inhibitors. Structure-based vs. diversity-based library design and high-throughput synthesis and screening Wyss, P. C., P. R. Gerber, et al. J Med Chem 46 2003
Case Study-3 : Aminothiazole Libraries¶
Design of Diverse and Focused Libraries¶
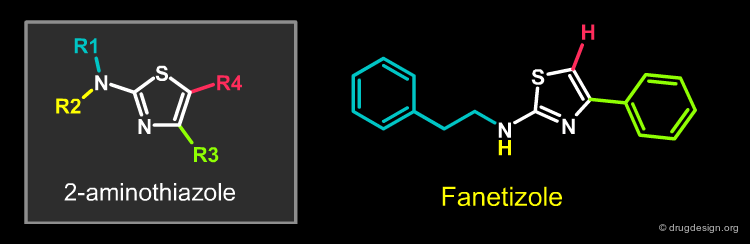
The design of a library of 2-aminothiazoles by a GSK team is presented here. A number of papers reporting molecules containing this structure have been published in recent years and show biological activities towards a diverse range of targets; this suggests that libraries of 2-aminothiazoles might have biologically active molecules of potential interest.
Author
Darren Green Director of Cheminformatics Science and Analysis, GlaxoSmithKline, Stevenage, UK
articles
A convenient procedure for the solution phase preparation of 2-aminothiazole combinatorial libraries N. Bailey, A.W. Dean, D.B. Judd, D. Middlemiss, R. Storer and S.P. Watson Biorg. Med. Chem. Lett. 6 (12) 1996
Steps in Library Design Process¶
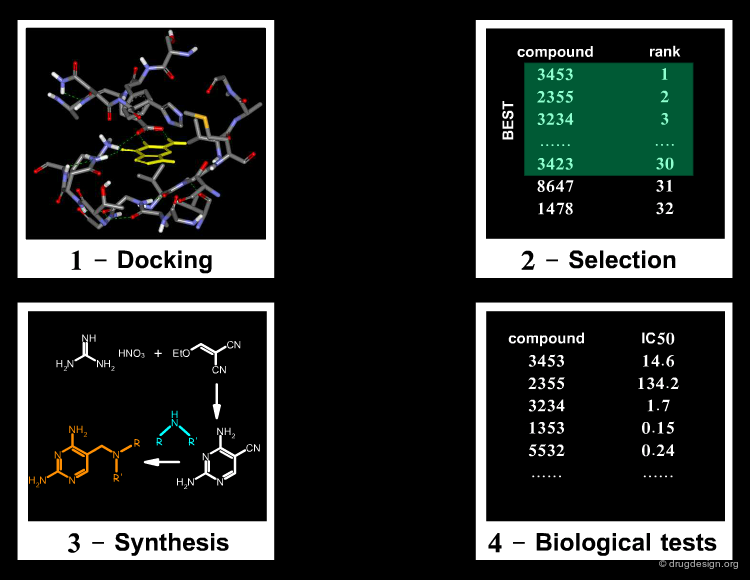
The typical steps involved in library design are shown in the figure below and will be illustrated for the design of the 2-aminothiazole library in the following pages.
Author
Darren Green Director of Cheminformatics Science and Analysis, GlaxoSmithKline, Stevenage, UK
articles
Implementation of a System for Reagent Selection and Library Enumeration, Profiling, and Design Andrew R. Leach, John Bradshaw, Darren V. S. Green, and Michael M. Hann J. Chem. Inf. Comput. Sci 39 1999
Define Chemical Reaction¶
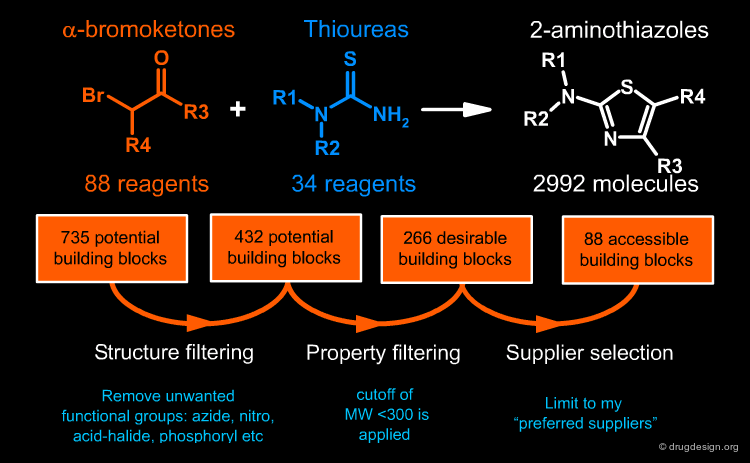
The first step is the choice of a synthetic scheme. In our example the Hantzsch synthesis was chosen. It is a robust and well established procedure for the preparation of 2-aminothiazoles from α-bromoketones and thioureas with a high yield and purity from readily accessible reagents. The reaction is simple, rapid, effective and enables the ready solution phase preparation of libraries of discrete 2-aminothiazoles.
Author
Darren Green Director of Cheminformatics Science and Analysis, GlaxoSmithKline, Stevenage, UK
articles
A convenient procedure for the solution phase preparation of 2-aminothiazole combinatorial libraries N. Bailey, A.W. Dean, D.B. Judd, D. Middlemiss, R. Storer and S.P. Watson Biorg. Med. Chem. Lett. 6 (12) 1996
Select Pool of Possible Building Blocks¶
The identification of possible reagents is done using databases of molecules. Traditional sources of reagents are databases such as the Available Chemicals Directory (ACD) from MDL. However, many suppliers now have their own websites, and there are now powerful web resources such as eMolecules. The identification of 735 potential reagents is shown below for the first building block, based on a substructure search using the query CO-C-Br.
Author
Darren Green Director of Cheminformatics Science and Analysis, GlaxoSmithKline, Stevenage, UK
Refine List of Building Blocks¶
Structure filtering is done to remove building blocks with unwanted functional groups using cheminformatics automated filtering tools. Then molecules with unwanted properties (e.g. high molecular weight) are removed and finally the selection is limited to preferred suppliers. A very important rule of library design is not to put any building block into the process that you are not happy to have selected. There is little point, and much frustration, in making a complex design only to find out that half of the selected building blocks are unavailable! The refinement process for the two building blocks led to a library of 2992 molecules.
Author
Darren Green Director of Cheminformatics Science and Analysis, GlaxoSmithKline, Stevenage, UK
Library Enumeration¶
The next step consists of the generation of the molecules of the virtual library in a computerized format (2D connectivity), a process called "library enumeration". This is a prerequisite for further analyses of the molecules, as will be presented later. There are several ways enumeration can be done, the two most popular being "reaction based" (transform-based enumeration) and fragment-based enumeration ("fragment marking").
Author
Darren Green Director of Cheminformatics Science and Analysis, GlaxoSmithKline, Stevenage, UK
Reaction-Based Enumeration¶
In reaction based methods, the chemical reaction transform is represented in a chemical language such as SMIRKS, and then the reagents are passed through the reaction. If the functional groups in the reagents match those in the reaction transform, then they "react" to produce the product, and any reagents that cannot react are eliminated.
Author
Darren Green Director of Cheminformatics Science and Analysis, GlaxoSmithKline, Stevenage, UK
Fragment-Based Enumeration¶
In fragment based methods, the chemist is asked to produce a core and R-group representation (familiar to anyone who carries out SAR analysis) by marking connection points onto the reagent structures. The core and reagents are then used by the enumeration program to generate the products.
Author
Darren Green Director of Cheminformatics Science and Analysis, GlaxoSmithKline, Stevenage, UK
Properties Profiling of the Virtual Library¶
Once the virtual library has been enumerated, a great number of properties can be calculated for each molecule of the library. This is done to select molecules that possess the desired profile whereas library members with undesirable properties are removed immediately.
Author
Darren Green Director of Cheminformatics Science and Analysis, GlaxoSmithKline, Stevenage, UK
articles
Implementation of a System for Reagent Selection and Library Enumeration, Profiling, and Design Andrew R. Leach, John Bradshaw, Darren V. S. Green, and Michael M. Hann J. Chem. Inf. Comput. Sci 39 1999
Simple Property Profiling¶
Simple properties such as logP, molecular weight etc... can influence the drug-likeness of a molecule. The following diagram illustrates the profile of our library of 2-aminothiazoles in terms of the cLogP (lipophilicity) distribution. It shows that about half of the compounds comply well with the Lipinski rule associated to LogP (cLogP less than 5). During our design phase, we will attempt to reduce the clogP of the compounds.
Author
Darren Green Director of Cheminformatics Science and Analysis, GlaxoSmithKline, Stevenage, UK
Profiling of Knowledge-Based Properties¶
More sophisticated properties can be predicted and profiled to help the chemist decide which molecules to synthesize. Examples of such properties include the affinity, pharmacokinetics and ADME/Tox properties of the molecules. Virtual screening incorporates knowledge derived either from structure-based or ligand-based information.
Author
Darren Green Director of Cheminformatics Science and Analysis, GlaxoSmithKline, Stevenage, UK
Analysis of the Diversity of the Virtual Library¶
An important element in the design of a virtual library is the selection of diverse molecules that best span the entire property space. One of the issues in library design is whether to consider the diversity of the building blocks (reagent-based diversity) or the resulting products (product-based diversity). The advantages and disadvantages of the two options are listed in the figure below.
Author
Darren Green Director of Cheminformatics Science and Analysis, GlaxoSmithKline, Stevenage, UK
articles
The effectiveness of reactant pools for generating structurally diverse combinatorial libraries Gillet VJ, Willett P and Bradshaw J, J. Chem. Inf. Comput. Sci. 37 1997
Optimal Subset of the Virtual Library for Synthesis¶
As the virtual library usually includes a huge number of compounds, what remains to be found is a method for reducing it to a practical size for synthesis. A judicious choice of building blocks will generate an optimal subset of molecules that have the desired properties.
Author
Darren Green Director of Cheminformatics Science and Analysis, GlaxoSmithKline, Stevenage, UK
Frequency Analysis Method¶
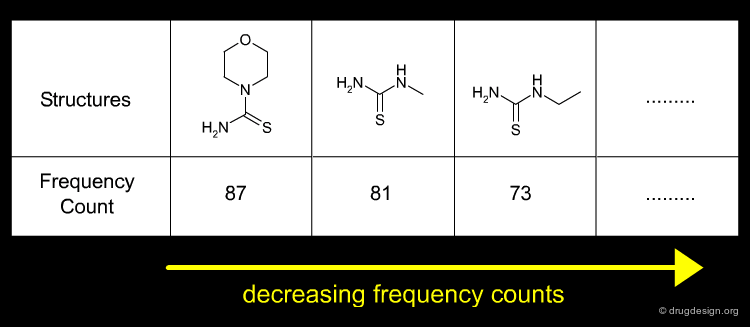
If we want to simply design an aminothiazole library with a drug like clogP distribution, there are some quick and effective techniques available, such as the Frequency Analysis introduced by Zheng. This approach consists of using a single filter (e.g. clogP<5) to identify "good" product structures. The number of times a building block appears in a "good" product is counted and those that appear most frequently are then selected. Results for the thiourea building blocks are shown below. The most frequent are small or hydrophilic building blocks, reflecting the selection criteria (clogP<5).
Author
Darren Green Director of Cheminformatics Science and Analysis, GlaxoSmithKline, Stevenage, UK
articles
Rational Combinatorial Library Design. 1. Focus-2D: A New Approach to the Design of Targeted Combinatorial Chemical Libraries Weifan Zheng, Sung Jin Cho, and Alexander Tropsha J. Chem. Inf. Comput. Sci. 38 1998
Advanced Frequency Analysis¶
Although frequency analysis works well on simple problems, it has some limitations. For example, if a high scoring building block only happens to combine well only with a set of low scoring building blocks which were not selected, then the method breaks down. Fortunately, frequency analysis can be enhanced by some simple iterations. This procedure can then be applied to analyze the best size for the library by monitoring its efficiency and effectiveness.
Author
Darren Green Director of Cheminformatics Science and Analysis, GlaxoSmithKline, Stevenage, UK
articles
PLUMS: a Program for the Rapid Optimization of Focused Libraries Gianpaolo Bravi, Darren V. S. Green, Michael M. Hann, and Andrew R. Leach J.Chem.Inf.Comput.Sci. 40 2000
Example of Advanced Frequency Analysis¶
Below is an example graph of Score (= efficiency + effectiveness; in yellow) versus library size as one building block is removed in turn, with the number of "good" molecules in the library shown in blue. The peak in the plot of the Score is often indicative of the best libraries to make; the analysis suggests here that the ideal library size is around 5000 compounds.
Author
Darren Green Director of Cheminformatics Science and Analysis, GlaxoSmithKline, Stevenage, UK
articles
PLUMS: a Program for the Rapid Optimization of Focused Libraries Gianpaolo Bravi, Darren V. S. Green, Michael M. Hann, and Andrew R. Leach J.Chem.Inf.Comput.Sci. 40 2000
Multicriteria Optimization¶
Library design is a multi-objective optimization problem. It is common to require a library design to meet many criteria: diverse, drug like, different to what you already have, similar to a lead molecule, predicted to be CNS penetrant, to have high bioavailablity and low plasma protein binding. There are a number of methods that aim to satisfy many criteria simultaneously. The weighted sum approach and the MOGA genetic algorithms are examples of such methods.
Author
Darren Green Director of Cheminformatics Science and Analysis, GlaxoSmithKline, Stevenage, UK
articles
Application of Nearest-Neighbor and Cluster Analyses in Pharmaceutical Lead Discovery David T. Stanton, Timothy W. Morris, Siddhartha Roychoudhury, and Christian N. Parker J.Chem.Inf.Comput.Sci. 39 1999
The Weighted Sum Approach¶
The first method to address the multiple criteria issue used what is known as a weighted sum approach; it is simple to understand and easy to implement. The weighted sum approach scalarizes a set of objectives into a single objective by multiplying each of them with a user-supplied weight. The weight of an objective is selected to be proportional to the objective's relative importance, and the global function to be minimized is a function of n.
Author
Darren Green Director of Cheminformatics Science and Analysis, GlaxoSmithKline, Stevenage, UK
articles
Current Status of Virtual Combinatorial Library Design Weber L QSAR and Combinatorial Science 24 2005
Limitation of the Weighted Sum Approach¶
The minimization of a weighted sum function yields only one solution, and this is an enormous drawback because this solution may be not optimal forany criteria. Averaging of too many objectives can lead to a solution which is uniformly average, and not optimal for any criteria. Moreover, in the weighted sum approach the combinatorics of the problem prevents us from studying every possible solution; for example, there are 1026 different ways of selecting 10 α-bromoketones and 10 thioureas from a 100x100 virtual library!
Author
Darren Green Director of Cheminformatics Science and Analysis, GlaxoSmithKline, Stevenage, UK
Multiple Objective Genetic Algorithms (MOGA)¶
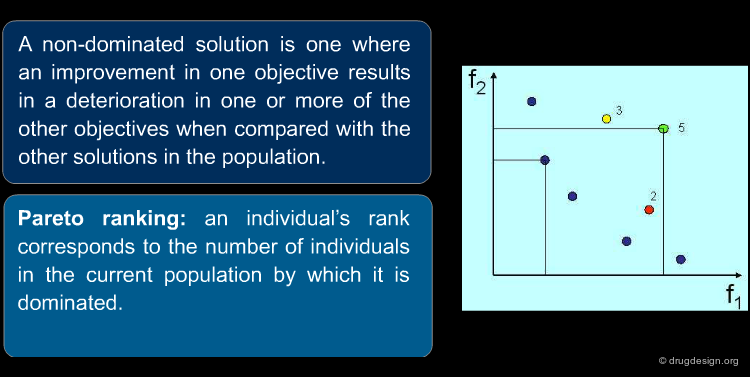
Genetic algorithms are easily adapted to use the Pareto method. These algorithms are known as MOGAs; they represent a powerful technique for multi-objective problems. The Pareto method uses ranking of solutions to enable comparison of non-commensurate data (cost, bioavailability) and provides multiple solutions (different compromises between the individual criteria) without having to weight the criteria a-priori.
Author
Darren Green Director of Cheminformatics Science and Analysis, GlaxoSmithKline, Stevenage, UK
articles
Combinatorial Library Design Using a Multiobjective Genetic Algorithm Valerie J. Gillet, Wael Khatib, Peter Willett, Peter J. Fleming, and Darren V. S. Green J.Chem.Inf.Comput.Sci. 42 2002
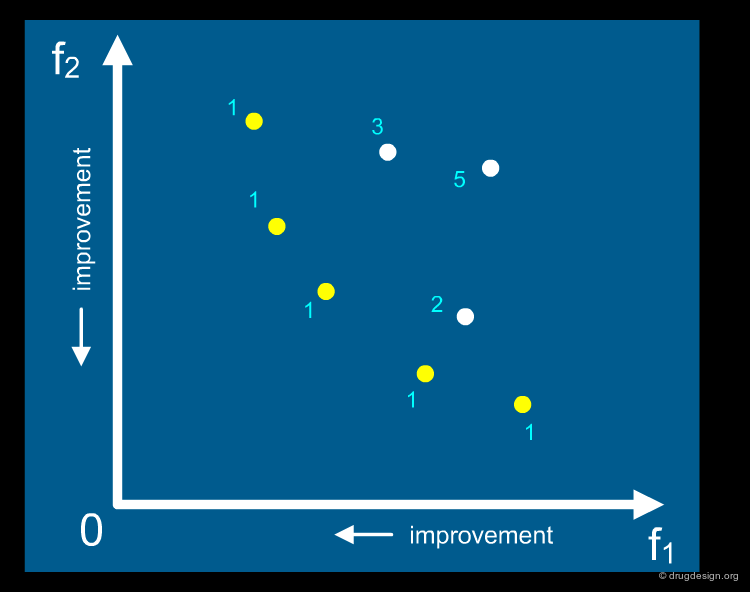
MOGA Plot and Pareto Ranking¶
An example of a MOGA result is illustrated below for a two-criteria optimization. Each point in the diagram corresponds to a solution. The Pareto method ranks each solution according to the number of individuals by which it is dominated (move the cursor on top of each solution to understand the ranking). The yellow points correspond to non-dominated solutions (optimal solutions); the library designer can chose from these different compromise solutions.
Author
Darren Green Director of Cheminformatics Science and Analysis, GlaxoSmithKline, Stevenage, UK
articles
Multiobjective optimization and multiple constraint handling with evolutionary algorithms. I. A unified formulation Carlos M. Fonseca and Peter J. Fleming IEEE Transactions on Systems, Man, and Cybernetics-Part A: Systems and Humans 28 1998 None
Example of Multi-Dimensional Optimization¶
The 2-aminothiazoles library design included the already known antifungal drug Fanetizole, a molecule with a high logP value. Applying the drug-like approach, the team searched in their library for a compound similar to Fanetizole but with a lower logP.
Author
Darren Green Director of Cheminformatics Science and Analysis, GlaxoSmithKline, Stevenage, UK
articles
A convenient procedure for the solution phase preparation of 2-aminothiazole combinatorial libraries N. Bailey, A.W. Dean, D.B. Judd, D. Middlemiss, R. Storer and S.P. Watson Biorg. Med. Chem. Lett. 6 (12) 1996
MOGA Results¶
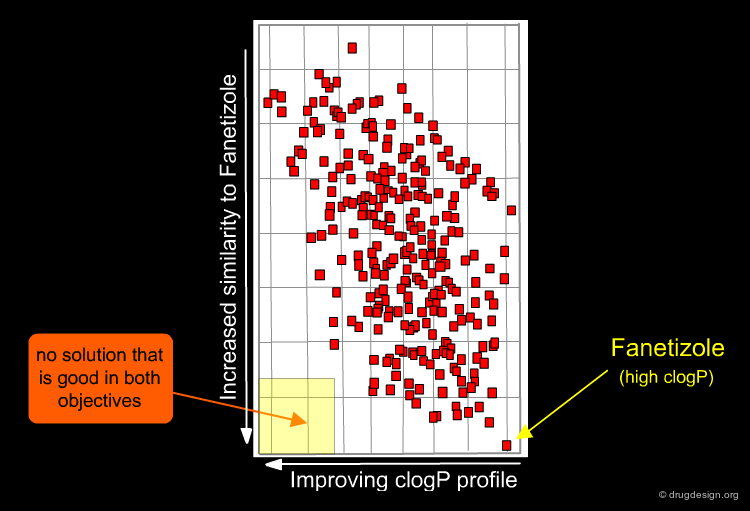
MOGA returns a family of solutions which are equivalent across the objectives that have been set; we now need to explore them. In the figure below, the solutions are shown in red, with their clogP, and similarity scores as compared to Fanetizole. It can be seen that these objectives compete: it is not possible to find a solution that satisfies both objectives.
Author
Darren Green Director of Cheminformatics Science and Analysis, GlaxoSmithKline, Stevenage, UK
Expanding one MOGA Solution¶
To find molecules with improved properties, the next step is to design a new focused library which is diverse with respect to Fanetizole and avoids the compounds with high clogP values. One solution from the MOGA diagram was expanded and four molecules resulting from this analysis are shown in the diagram below.
Author
Darren Green Director of Cheminformatics Science and Analysis, GlaxoSmithKline, Stevenage, UK
ADDITIONAL CASE STUDIES¶
Additional Case Studies¶
Copyright © 2024 drugdesign.org