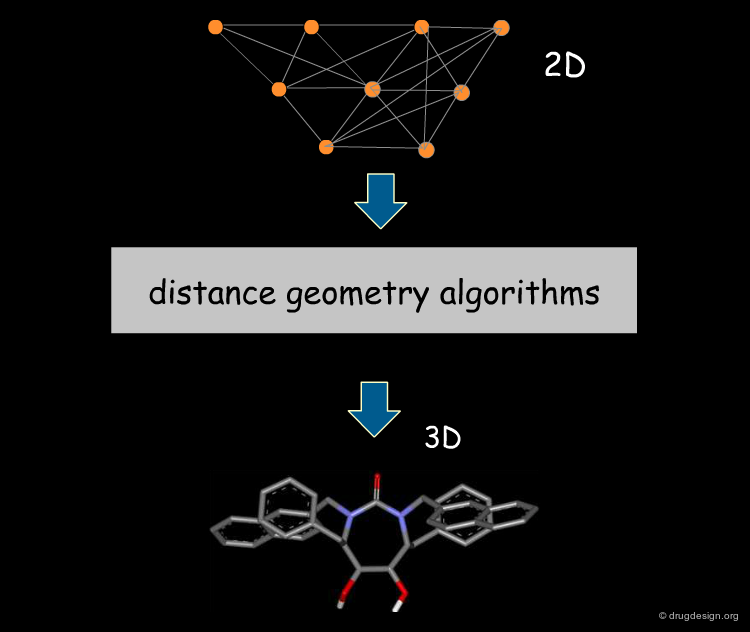
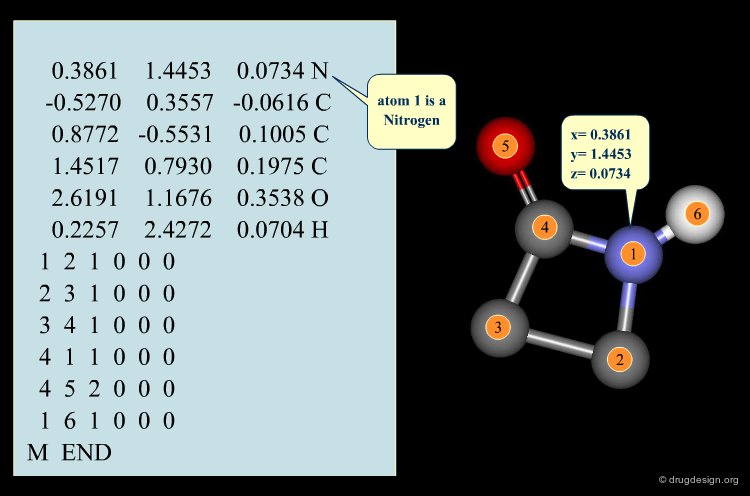
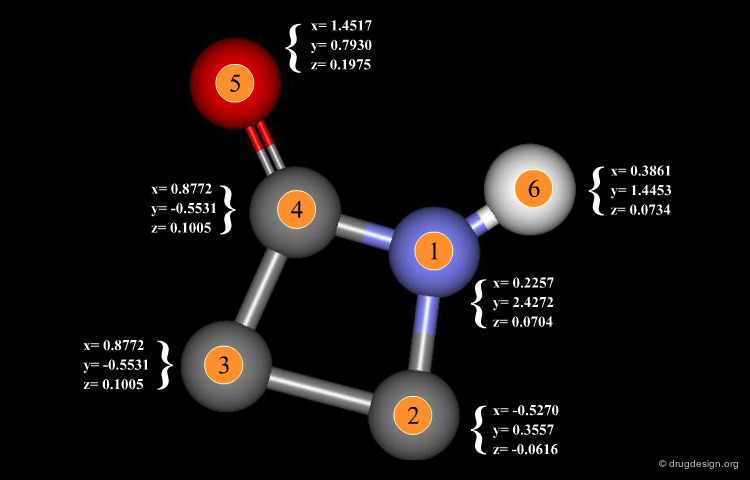
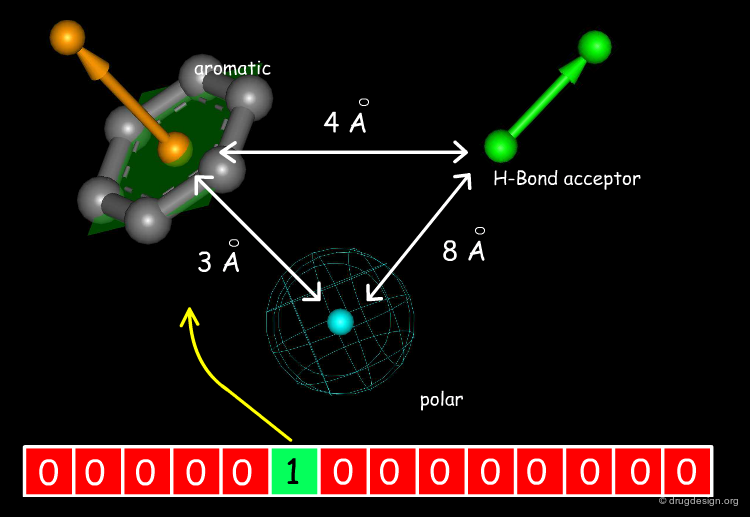
3D Database Searching¶
Info
3D searching is a technique that is based on searching in a database of molecules for compounds that contain structural elements as defined by a 3D query. This approach can be used for a wide variety of purposes ranging from testing a pharmacophore hypothesis to the design of a library for high throughput screening. This chapter is an introduction to this field and includes the following topics: the typical uses of 3D searching, the different types of searching, the construction of 3D databases and programs for 3D database searching.
Number of Pages: 63 (±1 hours read)
Last Modified: October 2004
Prerequisites: None
What is 3D Searching?¶
Importance of the 3D¶
The 3D molecular geometries are of major importance in drug design because they represent the very molecular determinants that control molecular interactions. They are essential to understanding and influencing biological processes and their possible aberrations.
What is 3D Searching?¶
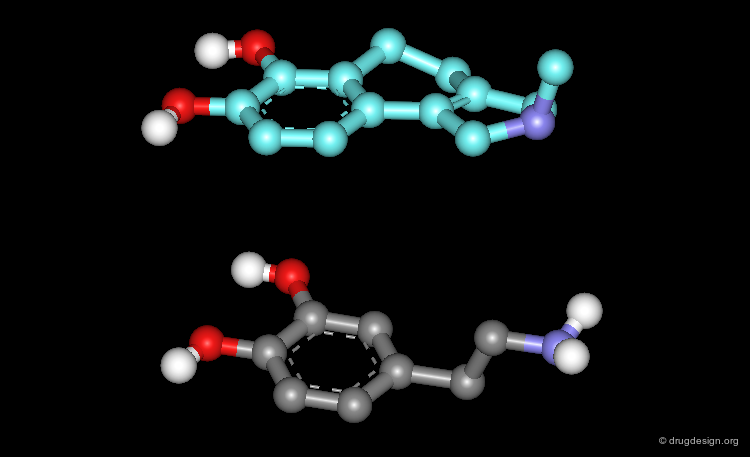
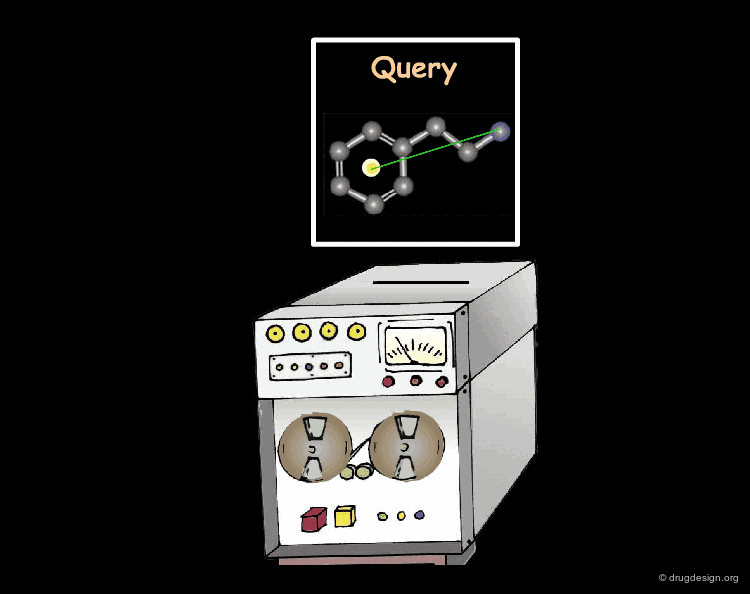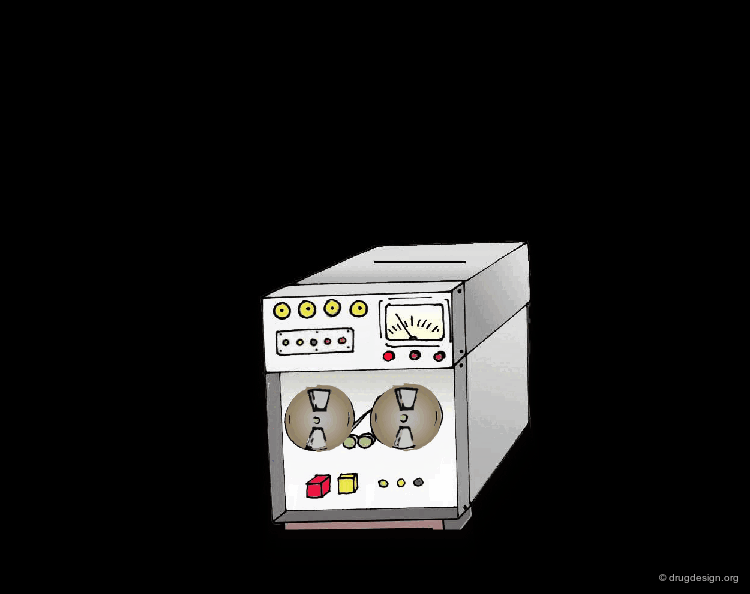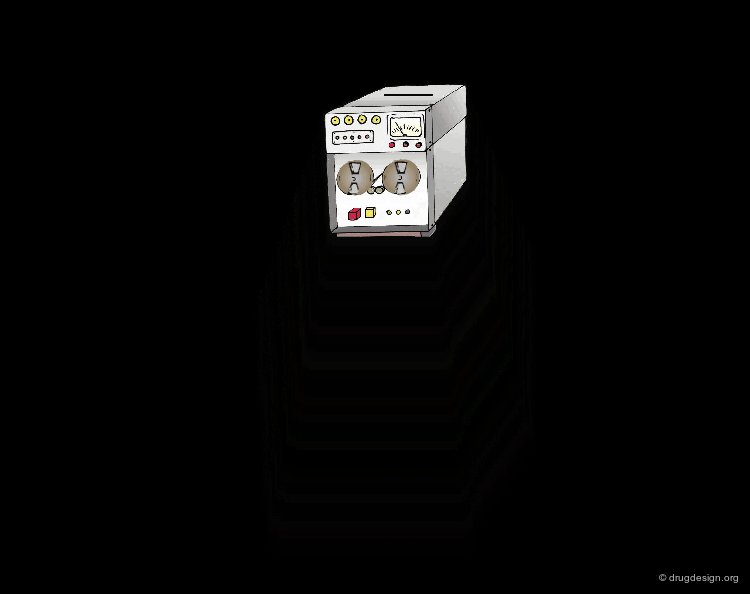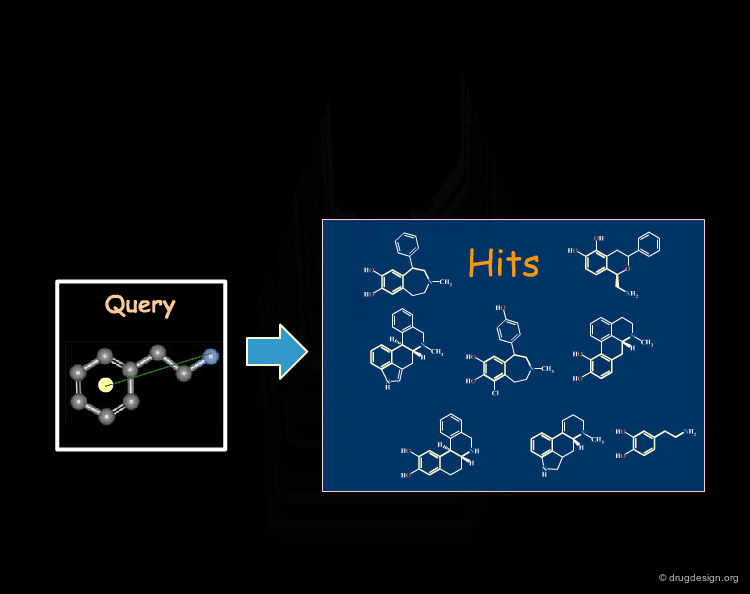
3D searching is a technique that is based on searching in a database of molecules for compounds that contain structural elements as defined by a 3D query. Potential bioactive molecules can be recognized, based on 3D properties. For example, the view shows a superposition of the dopamine query (grey) with a rigid analogue hit (blue) that is a potent D1 dopamine agonist.
Components of a 3D Searching Program¶
The components of a 3D searching program are as follows: (1) a database to search; (2) a search hypothesis; (3) a query; (4) mechanisms to enter the query; (5) computational mechanisms to apply the query to the database molecules along with a mechanism to limit the search by other user-supplied criteria; (6) mechanisms to display the hits in the context of the search query and (7) possibly a ranking of the hits.
3D Database¶
A 3D database is a collection of molecules organized so that it can easily be accessed, managed, and updated. A database can contain millions of compounds. In another section of this chapter we discuss the type of information that is stored in a 3D database and how a typical 3D database is constructed.
Search Hypothesis¶
A search hypothesis consists of 3D structural features that are desired to be found in molecules of the database. Composing this abstraction is often done by inspecting reference molecules in 3D. Sometimes, however, a preceding analysis will prepare the search query. The Catalyst program from Accelrys and the combination of DISCO and Unity from Tripos are examples of automatic preparation of the search query.
Converting a Search Hypothesis into a Query¶
Converting a search hypothesis into a 3D query to be understood by the program is generally easy to do. Most 3D searching programs include tools for describing the desired 3D features, qualitatively and quantitatively. A query can be an exact molecule or a fragment of a reference compound (direct query), or the result of a more subtle mental analysis (creative query).
Processing the Query¶
Processing the query consists of using it as input in the searching program to find out whether there are molecules (hits) that conform to the desired 3D requirements. The query is described with precise geometrical parameters.
Typical Uses of 3D Searching¶
Typical Uses of 3D Searching¶
3D searching can be used for a wide variety of different purposes, as outlined below. In the following pages we present and briefly discuss current most typical uses of 3D searching.
Test a Pharmacophore Hypothesis¶
3D searching is a logical method to validate a pharmacophore hypothesis. Typically a pharmacophore is expressed as distances between key points in a molecule, and the search query can be applied to a database to identify other molecules having the pharmacophore considered and test them biologically, to assess the validity of the hypothesis.
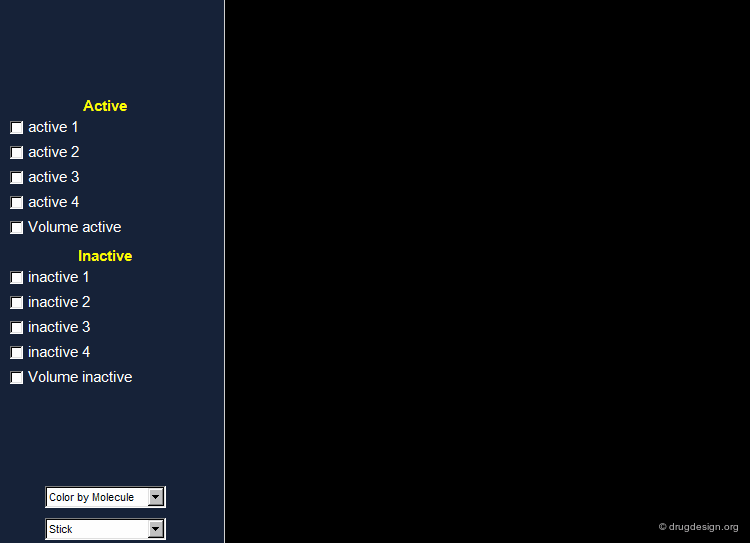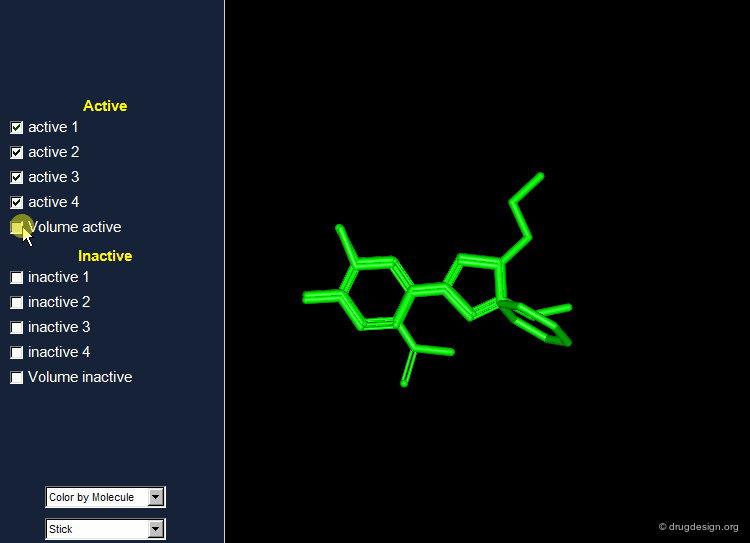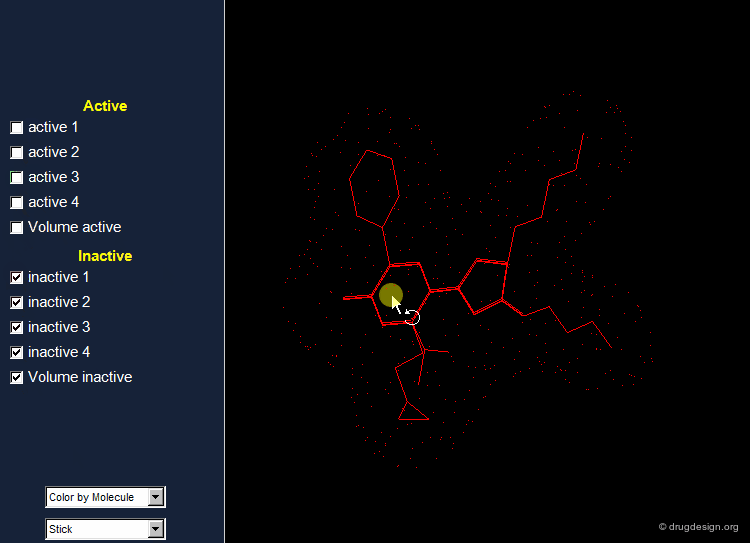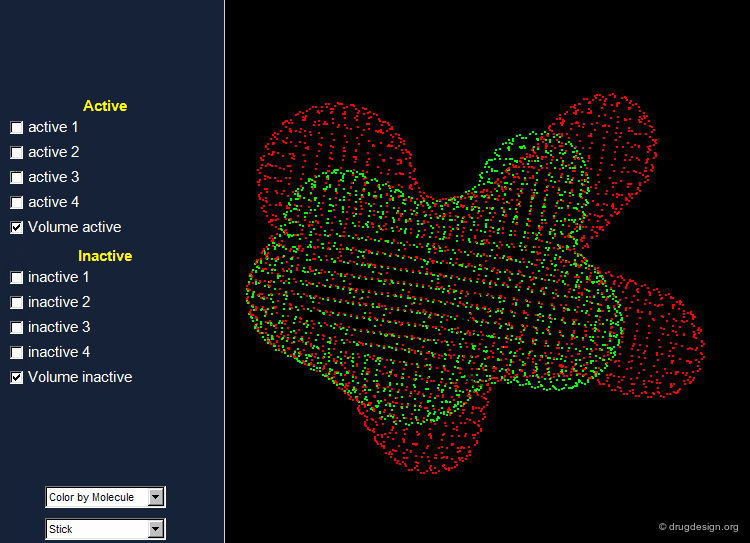
Find Hits that Fit the Volume of Active Molecules¶
A possible strategy in lead discovery is to search for molecules that fit the common volume of active compounds. For this purpose the set of all known active and inactive compounds are superimposed, and the resulting information is used for subsequent 3D searching.
Bioactive Conformation of a Flexible Molecule¶
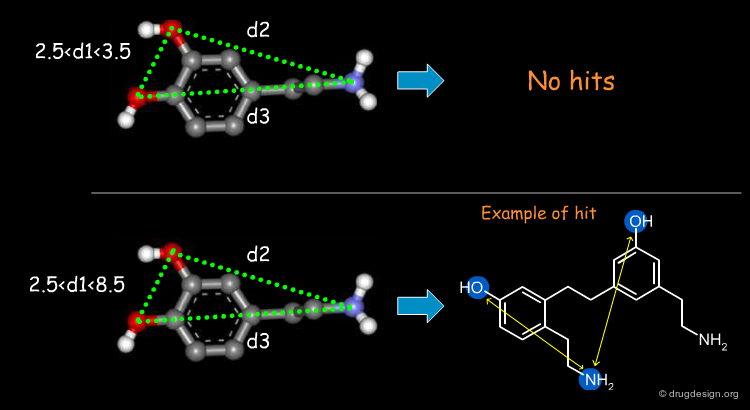
The first step in a drug design project might be to synthesize molecules that probe the potential bioactive conformation considered. For dopamine for example, 3D searching can be used to identify rigid analogue hits corresponding to different hypotheses (e.g. "Extended" or "Folded"). The biological testing of the hits obtained will indicate what is the conformation of dopamine when it binds to the receptor considered.
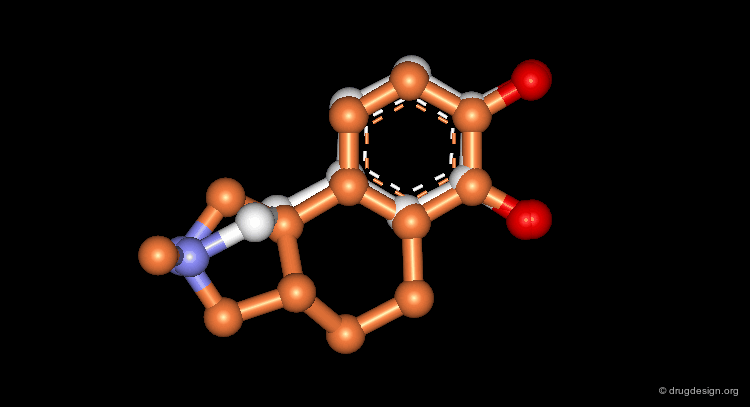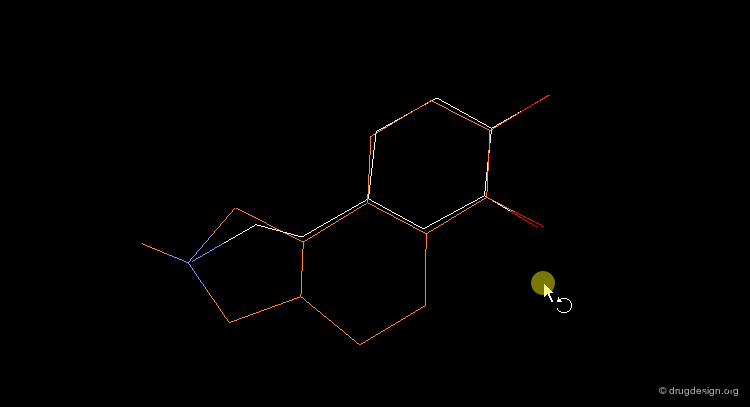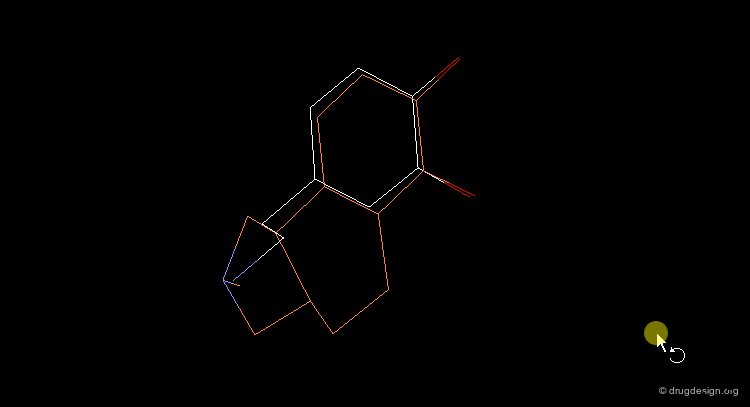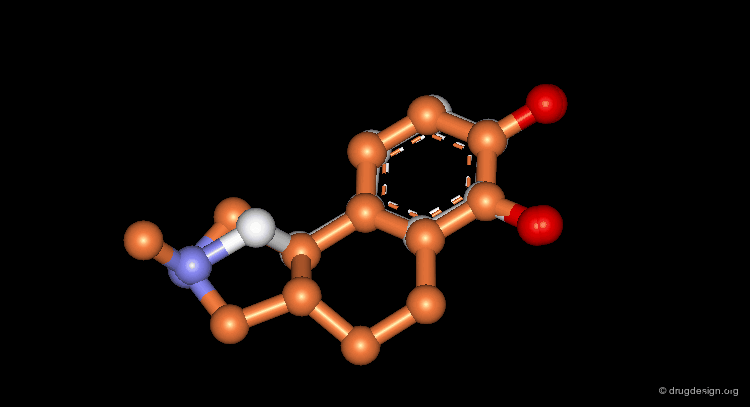
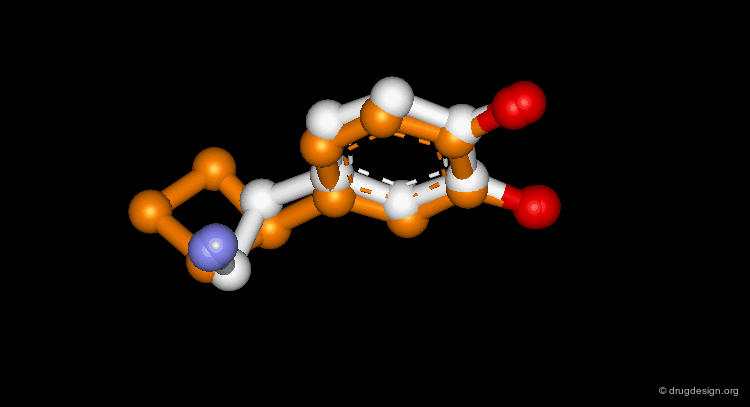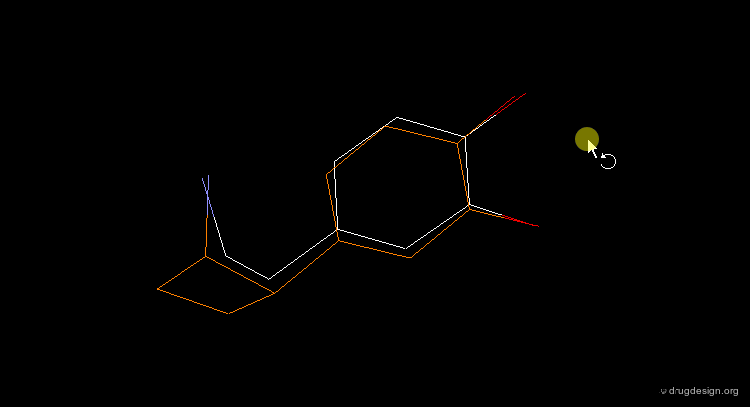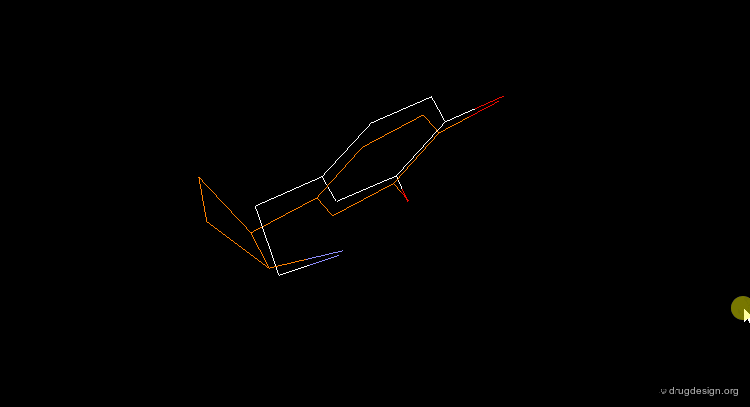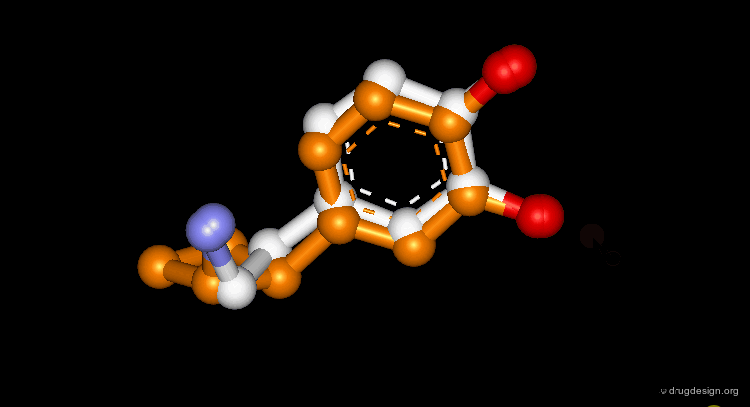
"Lead-Hop" to a New Core¶
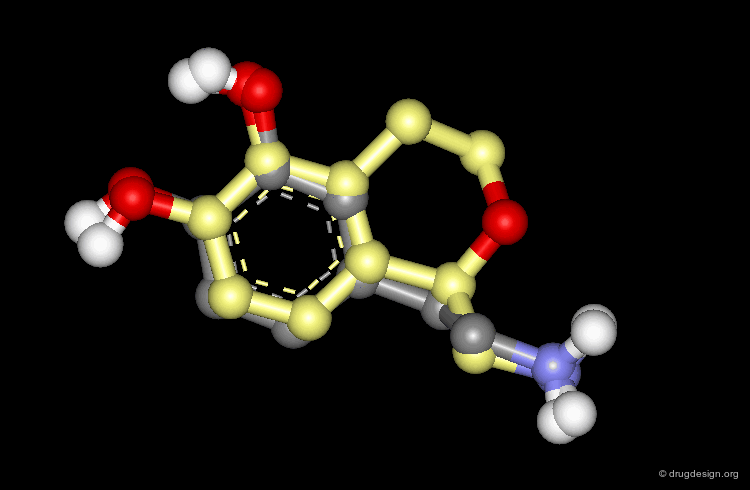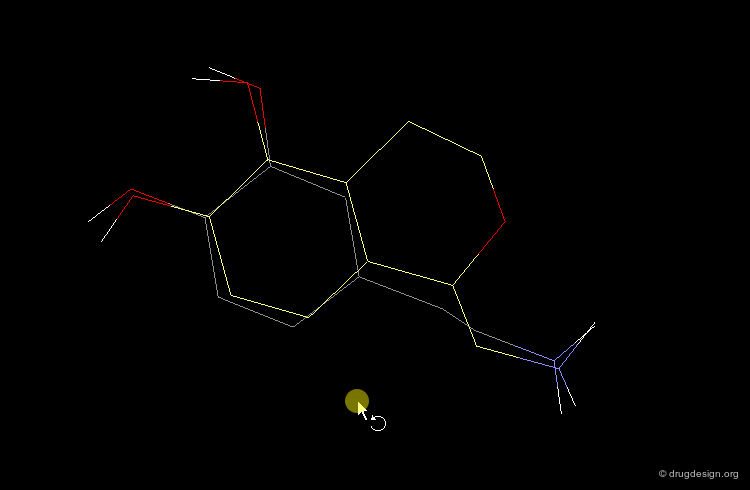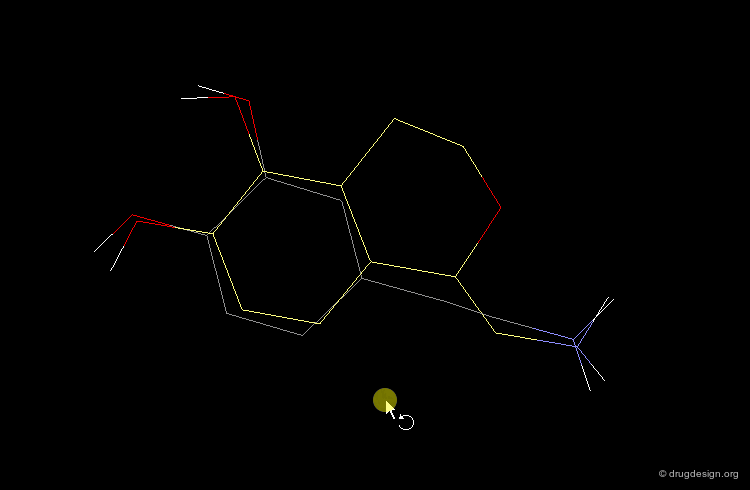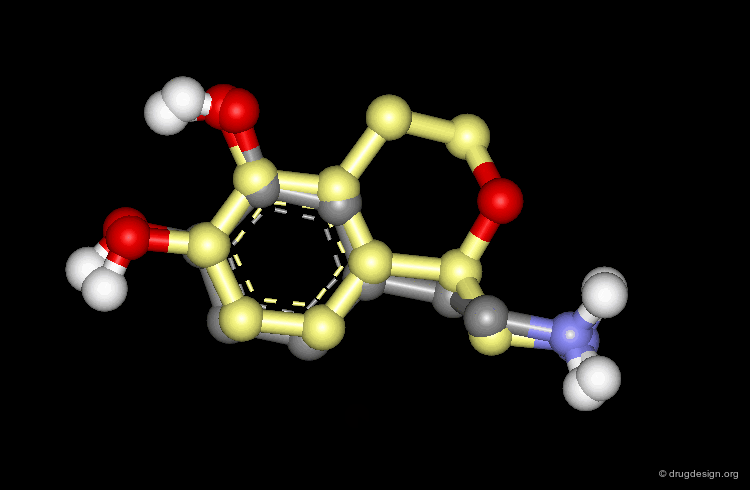
Once a bioactive conformation is established, one may wish to design one or more novel series that retain the target bioactivity. 3D searching can be used to "lead-hop" and find a new core. Shown in yellow is a novel D1 dopaminergic agonist that was found at Abbott, using the dopamine query in the extended conformation visualized below.
Design a Library for HTS¶
One way to test a pharmacophore hypothesis is to run a high throughput screen on additional compounds identified by 3D searching to fit the hypothesis. One could also use the results of multiple 3D searches to prepare a library that examines several competing pharmacophore hypotheses. Alternatively, the library designed from 3D searching might fulfill a pharmacophore common to several related biological targets (for example kinase inhibitors). In this case the library would be used to screen new targets for this biological class.
Structural Analyses on Experimental 3D Databases¶
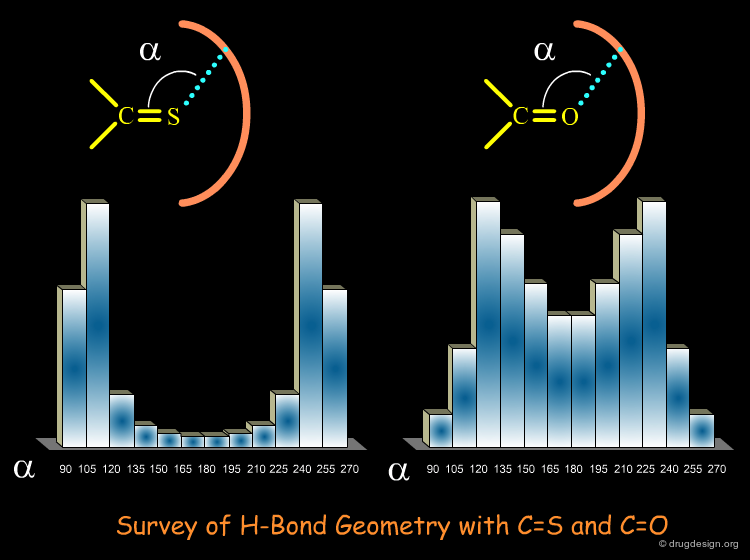
When the database contains experimental structures (for example from X-ray crystallography) it can be used to answer questions such as: what is the preferred orientation of two interacting phenyl rings; what are the H-bond angles and distances for C=S and C=O? what are the preferred conformations of a 12-membered macrocycle? what is the geometry of phenyl-sulfonamides? what torsion angles are found in β-turns etc... it can also be exploited to design and calibrate force-field components for specific structural moieties.
articles
The Hydrogen Bond in the Solid State Thomas Steiner Angew. Chem. Int. Ed. 41 2002
Non-hydrogen Bond Interactions Involving the Methionine Sulfur Atom Debnath Pal et al. Journal of Biomolecular Structure and Dynamics 19 2001
Cis Peptide Bonds in Proteins: Residues Involved,their Conformations, Interactions and Locations Debnath Pal et al. J. Mol. Biol. 294 1999
Generate Ideas¶
3D database searching is a method of choice for generating new ideas. The hits do not need to be available in the stockroom their role is only to provide new ideas. For example, to connect fragments in a pre-determined orientation, to create original scaffolds, to replace a template by a moiety with a simple synthetic route etc... In the example below a hit prompted the idea of considering ring moieties. By itself the hit was improper but helped to conceive a novel lead structure.
Types of 3D Searches¶
Types of 3D Searches¶
As the practice of molecular modeling has matured, variations of 3D searching have been implemented. The two main types of searches are geometric, which is based on distances and angles between points in the molecule; and shape, which matches the shape of a database molecule with that of the search query.
Geometric Searching¶
Geometric searching arose directly from the pharmacophore hypothesis and from the need to synthesize molecules to probe the bioactive conformation of flexible molecules. It defines the search by the geometric relationships between points, lines, and planes identified in molecules.
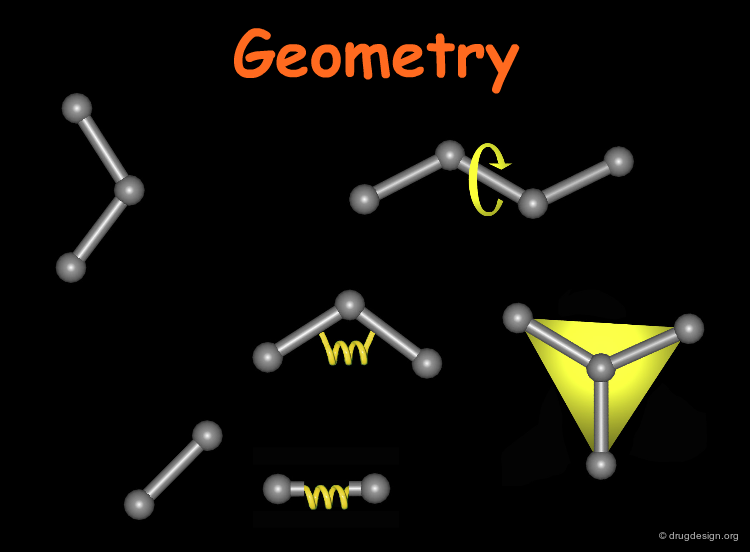
Typical Geometric Objects¶
To do a 3D search, the search question needs to be composed in a way that the 3D searching program can process. For geometric searching this involves specifying (1) how to use the database molecule to identify 3D objects such as points, lines, angles, and planes and (2) the 3D relationship between these 3D objects.
Typical Properties Considered¶
Every functional group has associated properties. The following are typically considered.
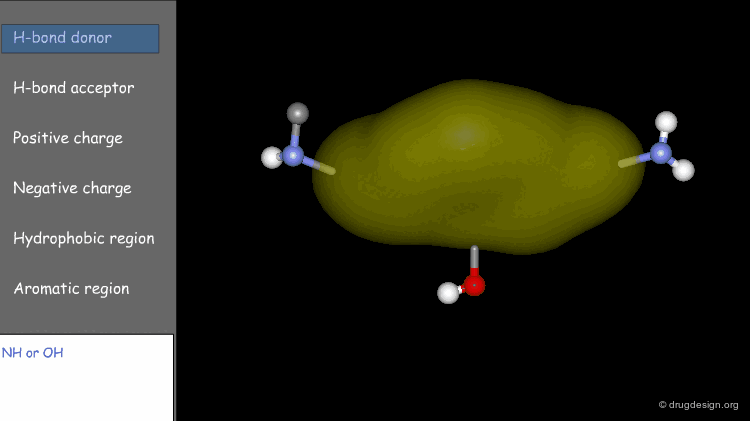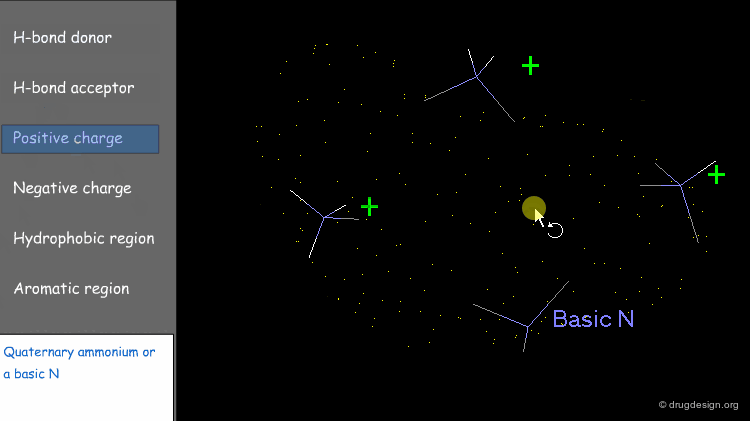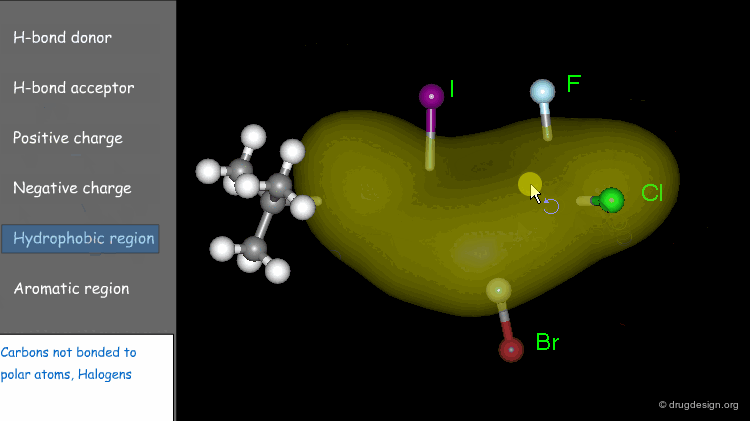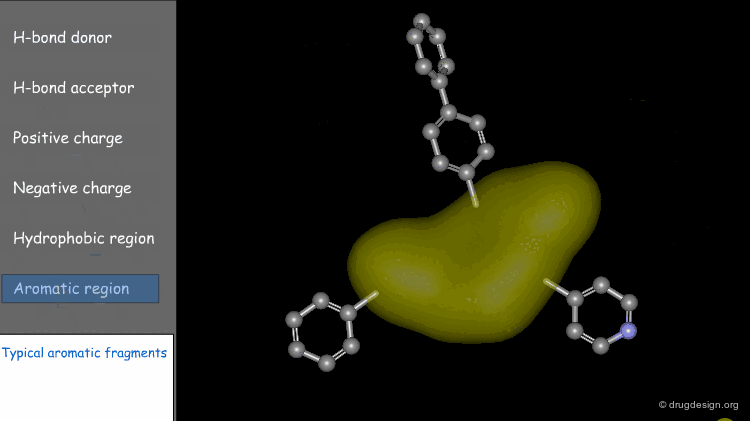
Point at an Atom¶
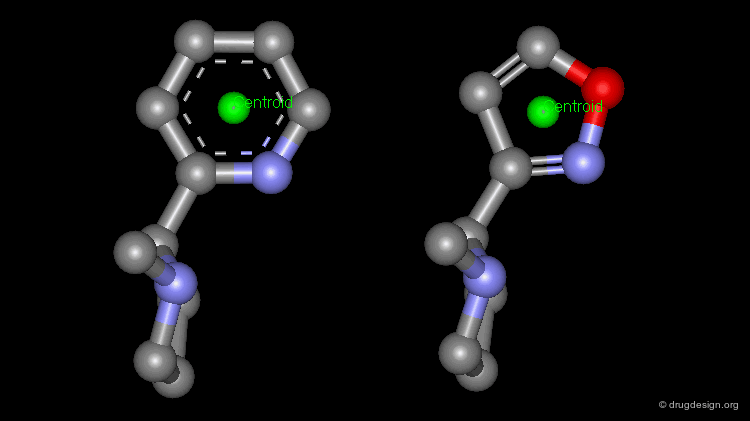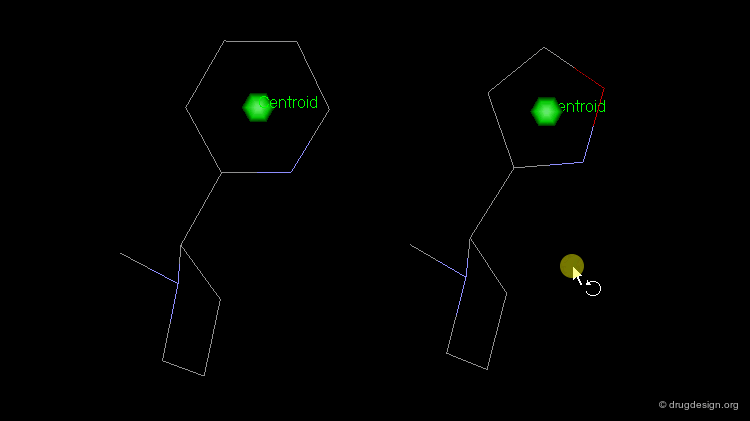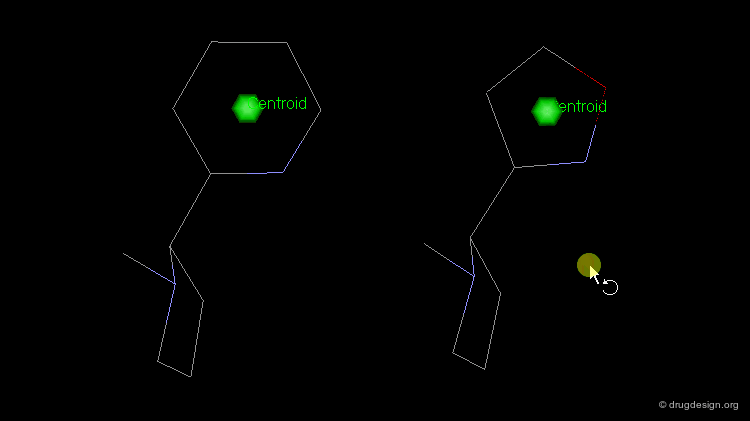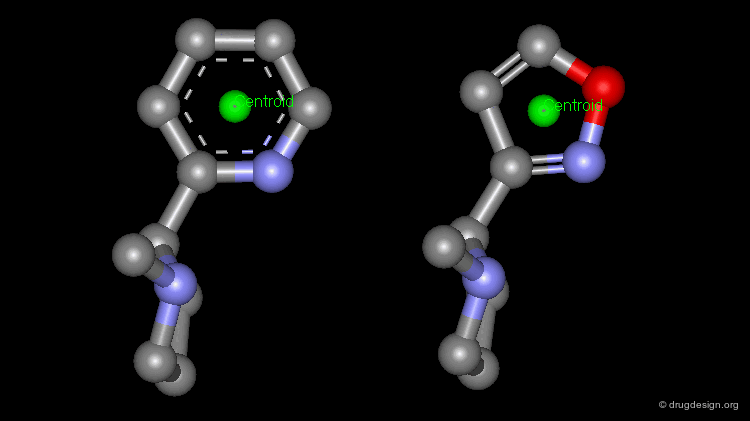
The simplest type of point is one centered at an atom. In this example H-bond donor points have been placed on the phenolic oxygen atoms and positive points on the basic nitrogen atom.
Centroid¶
Frequently the center of an aromatic ring is used as a point in the description. This makes a phenyl and a pyridine or an isoxazole equivalent.
Extension Point¶
To consider H-bonding groups, one extends along the line that describes the H-bond to the point at which the complementary atom on the protein would be, the site point. Shown are site points for nicotine (left) and dopamine (right).
Line¶
Sometimes a line is needed to orient the molecule with respect to the rest of the query. A normal (perpendicular) to a plane or another line can also be used to define a line.
Plane¶
A plane is typically defined by three atoms. To define the position of an aromatic system one typically defines a plane of the aromatic atoms and the centroid. However, other types of planes are equally useful, for example, the plane of the atoms in an amide.
Geometric Constraints¶
Geometric constraints describe the relationships between the geometric objects. They are typically given with a tolerance that describes the uncertainty of the value.
Distance Constraints¶
Most geometric searches include at least one distance constraint. If more than one distance to a particular point is used in a query, one must be certain to include enough distances to ensure that the hits meet expectations.
Angle Constraints¶
Frequently geometric searches include angle constraints. For example, in the query shown below angle constraints were incorporated, because the strength of the hydrogen bonds is dependent on the angle between the interacting groups.
Torsion Angle Constraints¶
A torsion angle describes the angle between two points at opposite ends of two other points. Typically this is the relationship between groups attached to opposite atoms of a single bond. To constrain the geometry either real or virtual torsion angles can be used (i.e. the four points are not connected by real chemical bonds and may also not correspond to real atoms).
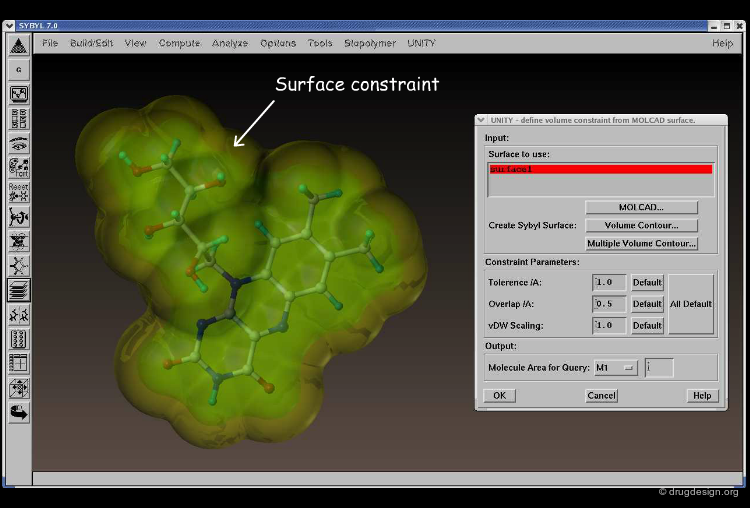
Excluded Volumes¶
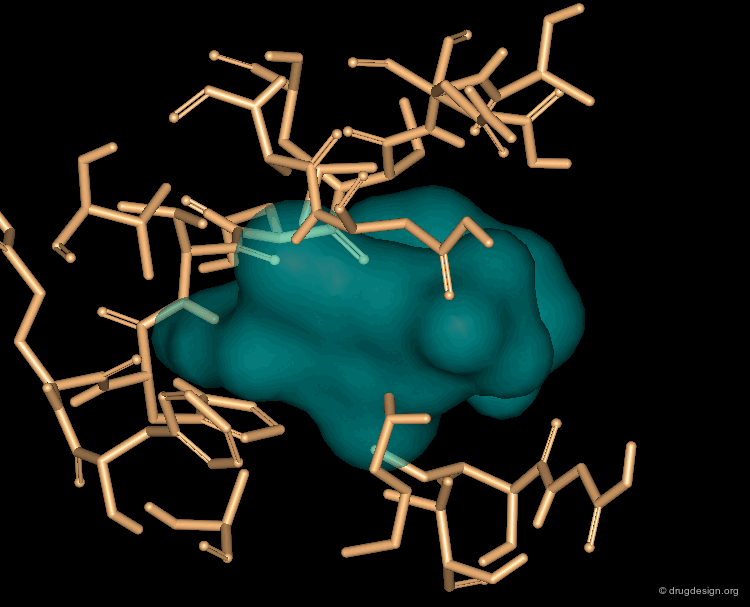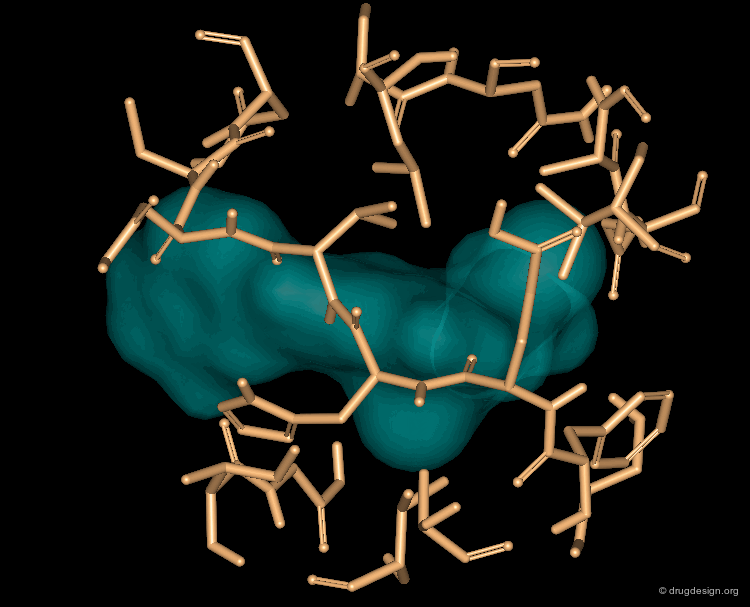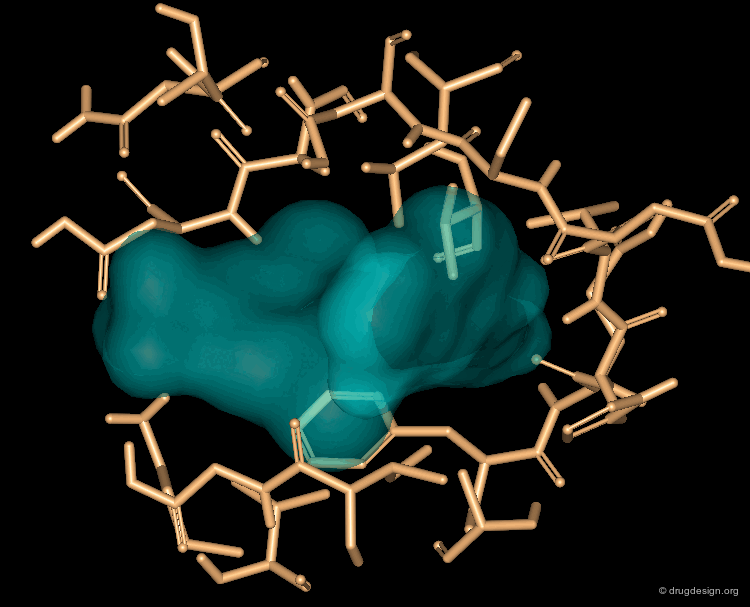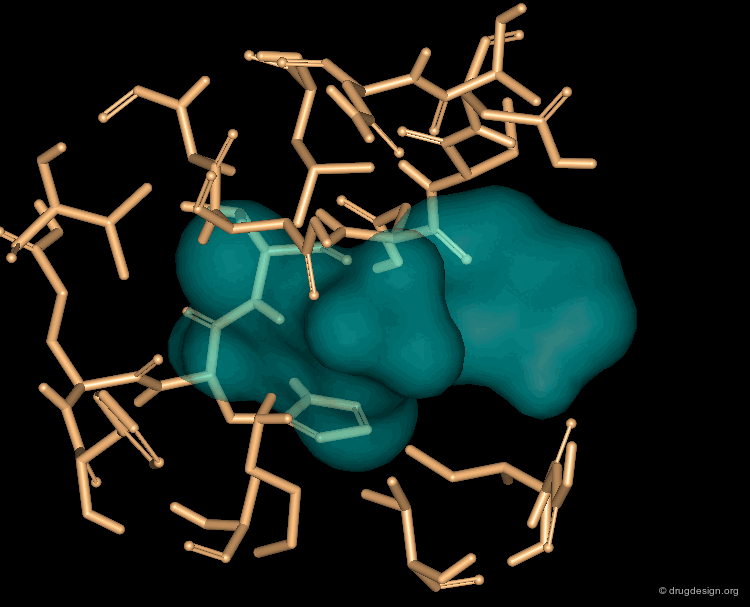
It is possible to use "excluded volume" constraints on a hypothesis to specify one or more regions that must not contain any atoms or bonds. Shown below, the use of an exclusion sphere, based on the observation that para-substituted analogs in this series are always inactive. The database search will therefore identify hits that have no atoms or bonds in this region.
Sources of Constraints for 3D Searching¶
Of course, one must have a basis for what the search is to find. Usually these constraints result from previous molecular modeling or experimental investigation of molecules of interest.
Constraints from a 3D Pharmacophore¶
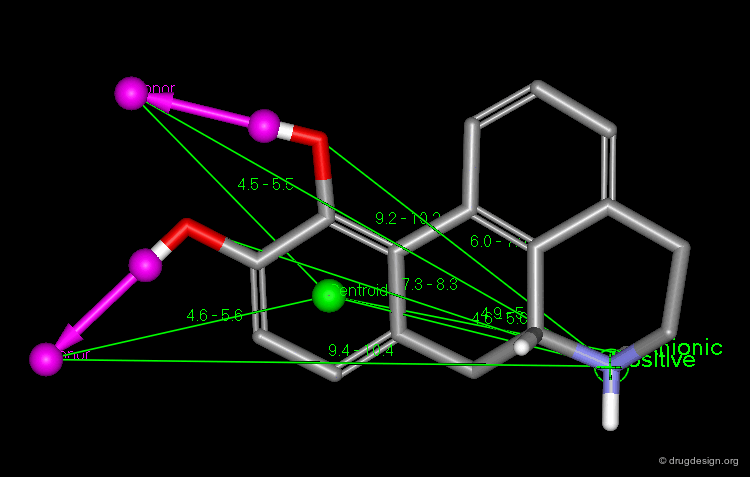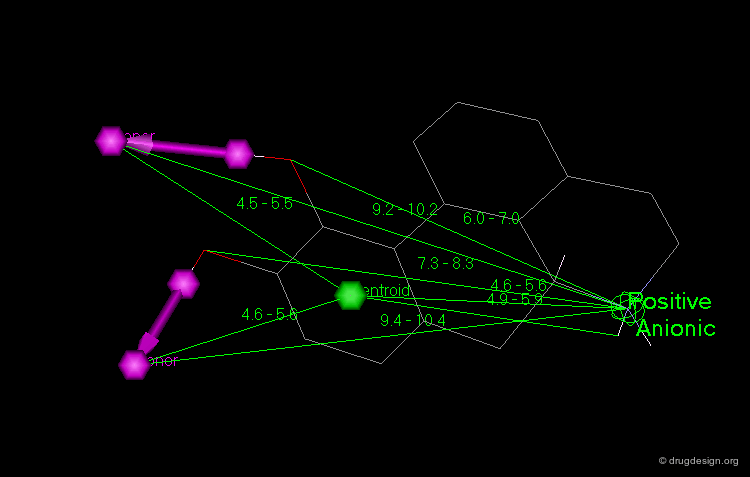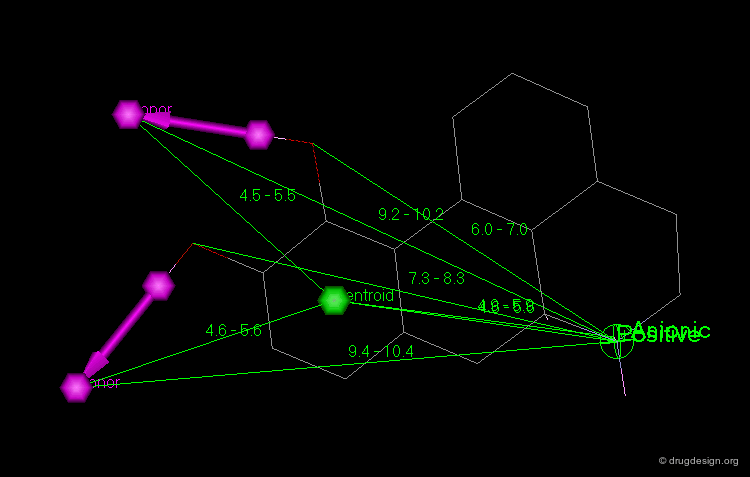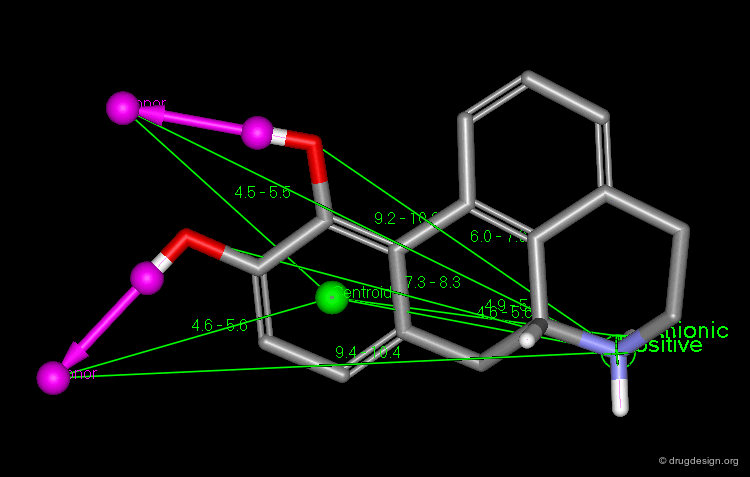
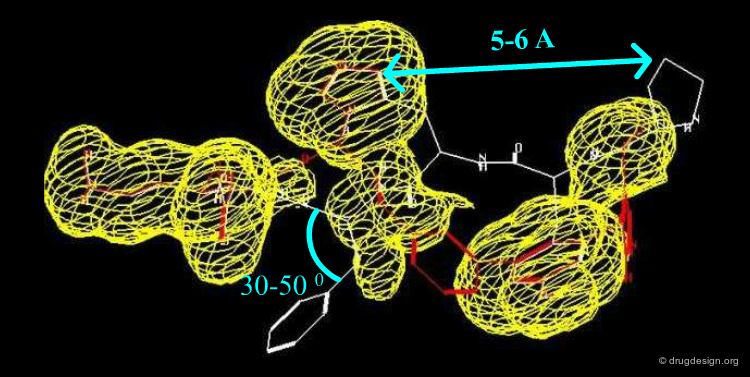
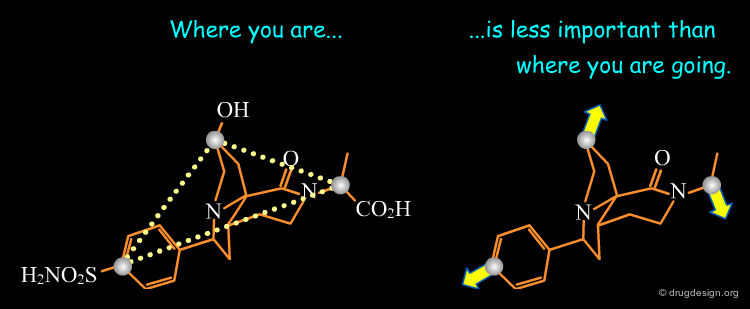
This diagram illustrates an example of constraints associated to a pharmacophore with a 3D query that has been used to identify new D1 dopamine agonists. Note the use of distances to a site point as well as to a phenolic oxygen.
Constraints from a Protein Active Site¶
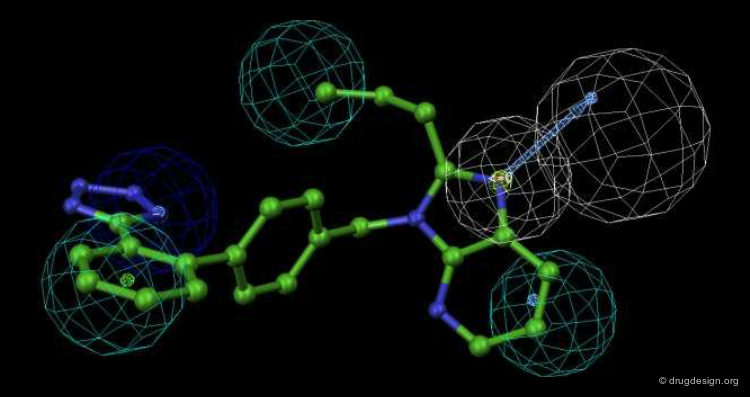
In this example the constraints were derived from the X-ray of a complex, and the query was designed to search for different cores that produce the same type of interactions. The diagram shows the H-bonding interactions of benzamidine with bovine pancreas beta trypsin. Both nitrogens form two close (<3.0 A) H-bonds to acceptor oxygen atoms in the protein.
Constraints from a Bioactive Conformation¶
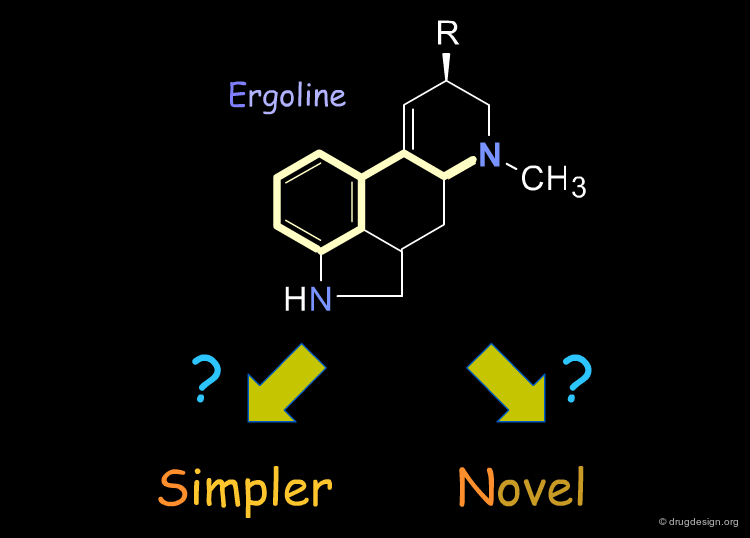
Given the known bioactive conformation of a molecule, one may wish to use 3D searching to find a simpler or novel compound.
Constraints to Probe the Bioactive Conformation¶
Constraints can be used to probe bioactive conformations. For example, the natural product apomorphine is a D1 and D2 dopaminergic agonist that constrains the geometry of the dopamine substructure. Because it is relatively rigid (except for inversion of the nitrogen), its bioactive conformation is known and can be used to set up a 3D search for novel dopaminergics. The constraints shown here are probably more complex than needed. One may find it preferable to try several searches.
Shape Searching¶
Geometric searching can include some implicit shape constraints by specifying distances that are not allowed. On the other hand, specific geometric searching programs can take the whole shape of the binding site into consideration. Docking programs do this explicitly. However, there are also programs that do not use the 3D structure of the target macromolecule as constraints for the search.
Bioactive Conformation as a Source of Shape¶
One could do a shape query on the molecule that defines the proposed bioactive conformation and/or search for molecules to test alternative proposals of the bioactive conformation.
Ensemble of Active Molecules as a Source of Shape¶
The shape of the space occupied by the ensemble of superimposed active molecules describes a powerful shape query.
Active Site as a Source of Shape¶
One can use the shape of a protein binding site as a shape query.
Full versus Partial Match¶
Any search implies that the database molecule must match all aspects of the search query. However, most programs allow you to specify that certain constraints and/or a certain percentage of the constraints need not be met by a hit.
Complex Query Combination¶
Although strict queries are generally used in 3D database searching, it is however possible to express a given query in terms of Boolean functionalities (such as AND, OR, NOT etc... and their combinations) to satisfy a logical combination of queries. This may be reserved for special situations or be used to reduce multiple searches to a single one. As in 2D searching, the search can be restricted by additional criteria, for example the molecular weight or the number of rotatable bonds etc... should not exceed specified values.
Constructing 3D Databases¶
Constructing 3D Databases¶
For 3D database searching one must decide the compounds to be searched, how the 3D conformations will be generated, what will be stored (and how) and the program that will be used for the searching. All of these components are discussed in the following pages.
Sources of Compounds for Searching¶
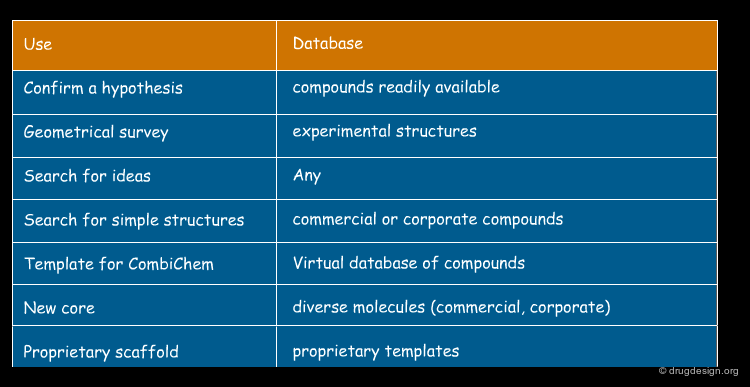
Because database preparation and screening is done on the computer, there is no need for the database to contain molecules that are in your stockroom, or indeed that are in anyone's stockroom. The source of compounds for the database depends on how you intend to use the results. Some typical uses of 3D database searching are indicated below.
Database of the Corporate Collection¶
If you work for a company and your objective is to select compounds for screening, then a good source of compounds for a 3D database is your corporate collection. Samples of many of these compounds are readily available and their biological properties will also be available. Note that these compounds might already have been tested for the activity of interest using high throughput screening!
Databases of Vendor Compounds¶
If you are looking for additional or different compounds to screen or to test your pharmacophore or bioactive conformation hypothesis, then a 3D database of vendor compounds is useful. Dozens of vendors of compounds for screening are happy to send CDs of their structures. One disadvantage of this source of compounds is that it takes time for the compounds to arrive and some will not be available. A second disadvantage is that typically anyone can purchase these compounds, hence your competitor might have already purchased samples of those that you identify.
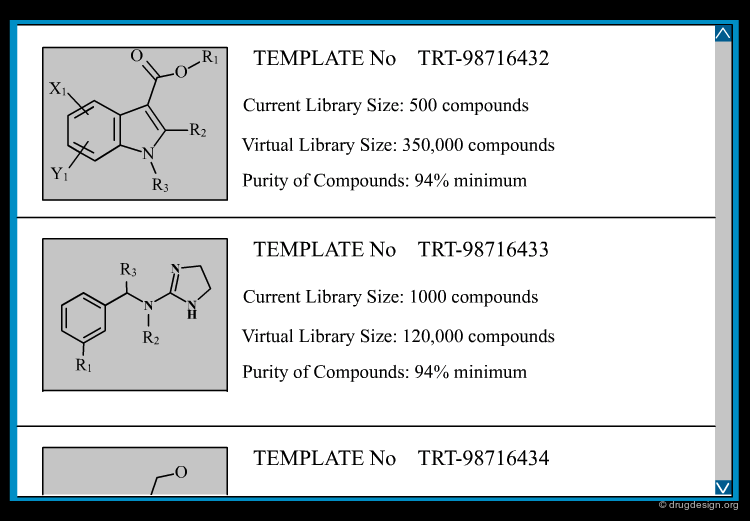
Database of Virtual Compounds¶
You might search a 3D database of virtual compounds to see which of those proposed for synthesis match a pharmacophore hypothesis. Alternatively you could generate and search one or more combinatorial libraries to have easily synthesized compounds available should you make a hit in a search. If the libraries are derived from proprietary chemistry, then one would expect a higher likelihood of patentability of any hits found active compared to hits found from vendor libraries.
Generating 3D Structures¶
Although some of the earliest 3D searching programs depended on carefully modeled 3D structures, these days the structures for searching are either determined crystallographically or generated by a fast 2D->3D structure converter program. Low energy conformers are collected and stored in the database however they cannot exceed a reasonable number because of limitations in storage capacity.
Experimental 3D Structures¶
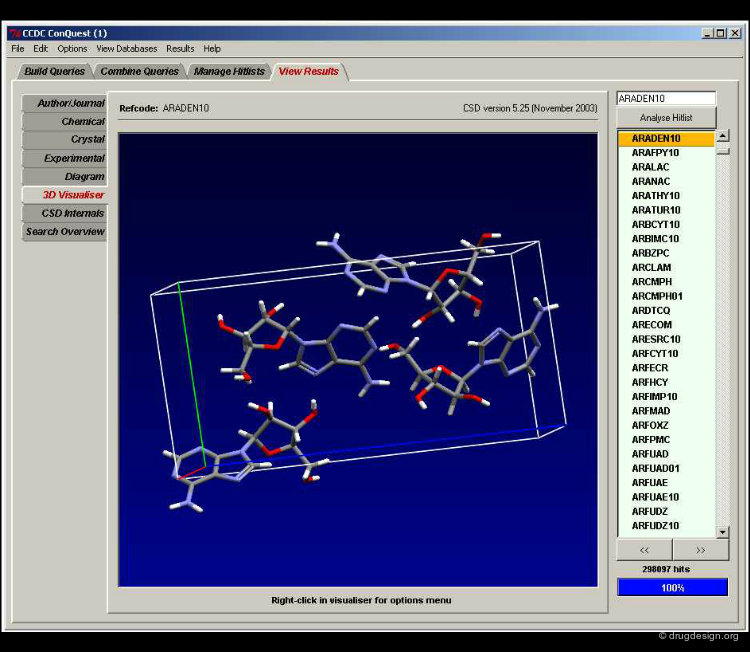
The Cambridge Structural Database contains the 3D coordinates of more than 260,000 compounds. Most of these were determined by X-ray diffraction, the remainder by neutron diffraction. The ConQuest software provides geometric 3D searching but also searching by name, substructure, and various crystallographic features.
Computationally Generated 3D Structures¶
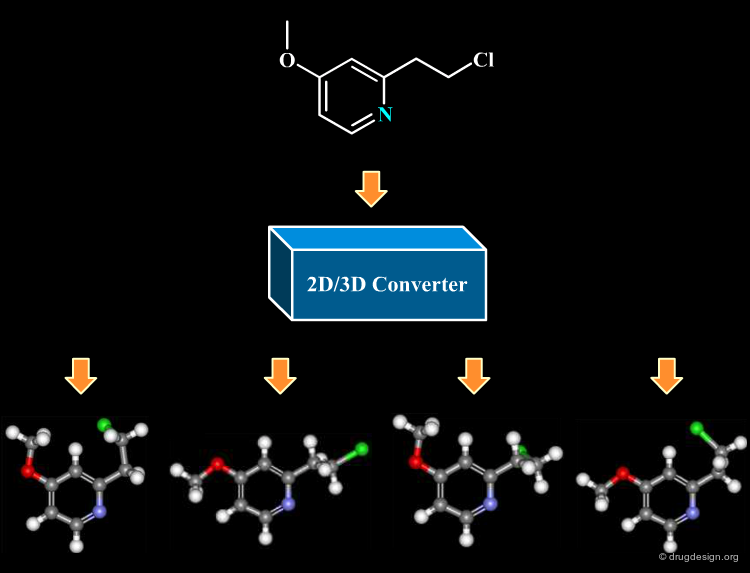
Converting the molecules contained in a database of compounds stored in 2D to 3D has become a routine operation. Many 2D->3D converter programs now exist; a brief outline of the most popular ones is given in the following pages.
Cleaning up 2D Input¶
Before generating a 3D structure it is important to "clean up" the 2D structures being used: remove any counter ions, salts, or water molecules that might be part of the registered structure. Remove any reactive or otherwise undesirable compounds. Possibly generate all stereo isomers, tautomers, and protonation states of the structures.
CONCORD¶
CONCORD (CONnections to COoRDinates) is a popular program for generating 3D databases from structure diagrams. These programs use a combination of rules and simple energy calculations to produce a 3D structure. The structures produced are typically very close to the one crystallographically observed, and usually are a very low energy, if not the minimum energy conformation. CONCORD generates 3D structures very quickly from all of the popular input formats.
CORINA¶
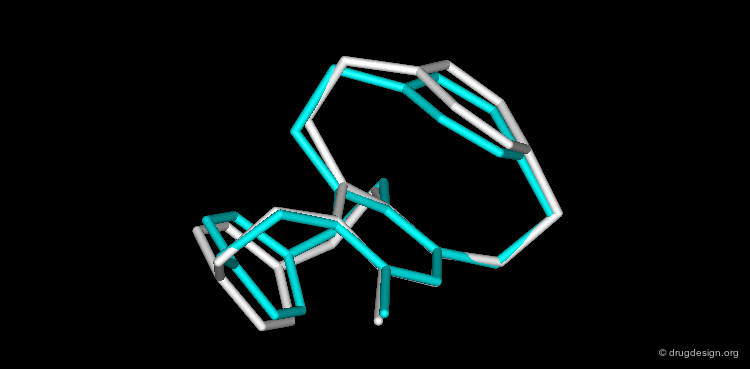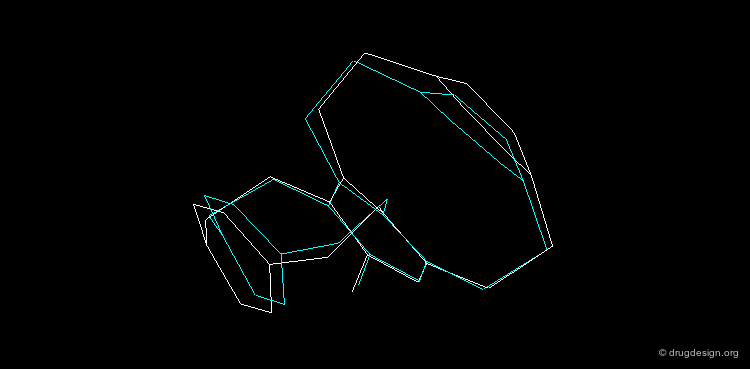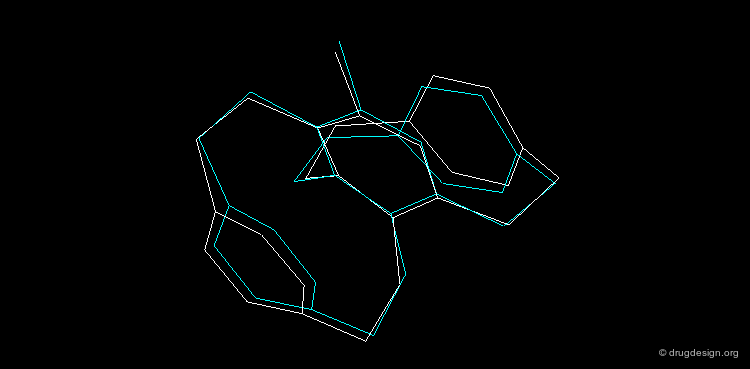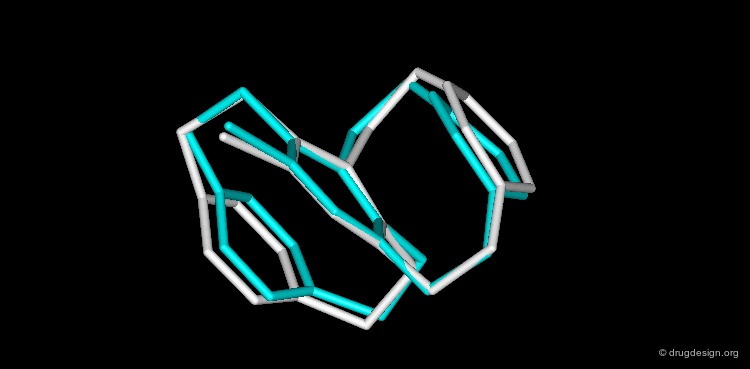
CORINA is a popular program that automatically generates three-dimensional atomic coordinates from the constitution of a molecule as expressed by a connection table or linear code. The program has proven to be reliable to convert large databases, for example a database with more than six million compounds has been converted with a conversion rate of 99%. The following view visualizes the comparison of the structure of cyclophane as generated by CORINA and experimentally determined by X-ray crystallography (RMS=0.26).
ConFirm and Omega¶
ConFirm and Omega generate and store many 3D conformations of a molecule for use in 3D database searching or for pharmacophore generation. The structures are generated by advanced distance geometry algorithms that eliminate duplicate conformations and attempt to cover all the conformational space.
Information Stored for a Molecule in the Database¶
The following information on a molecule is stored in the database: its 2D structure diagram, the coordinates and atom type of each atom in the conformation, the correspondence between the atoms in the 2D and 3D structures, features of the 2D and 3D structures that allow the program to filter out structures that cannot match the search query. The 3D filter can be as simple as the maximum distance between any pair of atoms or as elaborate as 3D fingerprints that encode the presence or absence of a particular distance/atom pair or triplet in the molecule.
Coordinates¶
The (X,Y,Z) coordinates of each atom in the conformation are stored in the database. They are directly taken from experimental results (X-rays) or generated by 2D-3D converter programs.
Bitmaps or Fingerprints of Typical Search Constraints¶
Chemical structures can be represented by "bitmaps" or "fingerprints", which are binary bit string representations of on/off values, each indicating the presence or absence of certain structural features in the molecule. The bipmap also includes 'digested' 3D information enabling rapid filtering of the desired 3D constraints.
Programs for 3D Searching¶
Programs for 3D Searching¶
Each program has its particular advantages and disadvantages. However, it is usually possible to use any of the programs described to identify the structures of interest. The programs are listed in the approximate order of their publication date.
ConQuest¶
ConQuest searches experimental 3D structures. This is an advantage because any retrieved structure will be a low energy conformation. This is a disadvantage if compounds of interest are not in the database. ConQuest can also be used to search for and/or visualize intermolecular contacts. This information can be helpful in generating a pharmacophore model 3D search question from compounds with unusual functional groups.
CAVEAT¶
CAVEAT is specifically designed to quickly search for templates that can constrain the conformation of important rotatable bonds of a flexible molecule. It works with sources of 3D structures such as the Cambridge Structural Database, to identify molecular skeletons that hold the functional groups in the desired 3D arrangement. CAVEAT is supplied with TRIAD and ILIAD, databases of hydrocarbon templates that contain 411,000 unique tricyclic structures and 110,000 acyclic frameworks. The particular molecules to be synthesized are derivatives of the templates in which carbons are replaced by heteroatoms for synthetic ease.
articles
CAVEAT: A Program to Facilitate the Design of Organic Molecules Lauri, G.; Bartlett, P.A. J. Comp. Aided Mol. Design 8 1994
UNITY¶
UNITY combines traditional 2D substructure with 3D searching in an environment tightly coupled with the Sybyl molecular modeling program, which provides pharmacophore perception and spreadsheet capabilities. A key feature of UNITY is that one need not store all conformations of a molecule in order to search them later. Rather, UNITY generates conformations that match the distance, angle, and shape constraints. It is able to do this quickly because when one loads a molecule into the database, 3D fingerprints are generated to encode its conformational possibilities.
articles
3D Database Searching in Drug Design Y.C. Martin J. Med. Chem 35 1992
ISIS¶
The standard ISIS structure searching tool also has capabilities for 3D searching in combination with 2D searching. One draws the query using ISIS-Draw, and no molecular modeling is required if one starts with a structure in an ISIS 3D database. The 3D structures in the databases provided by MDL are generated with CORINA.
Catalyst¶
Catalyst provides an integrated suite of software to process a set of active molecules, define a pharmacophore/3D-QSAR hypothesis, and/or search databases for molecules that match the hypothesis or that fit into a particular shape. Catalyst databases are prepared with many low energy 3D conformations of each molecule.
FlexS¶
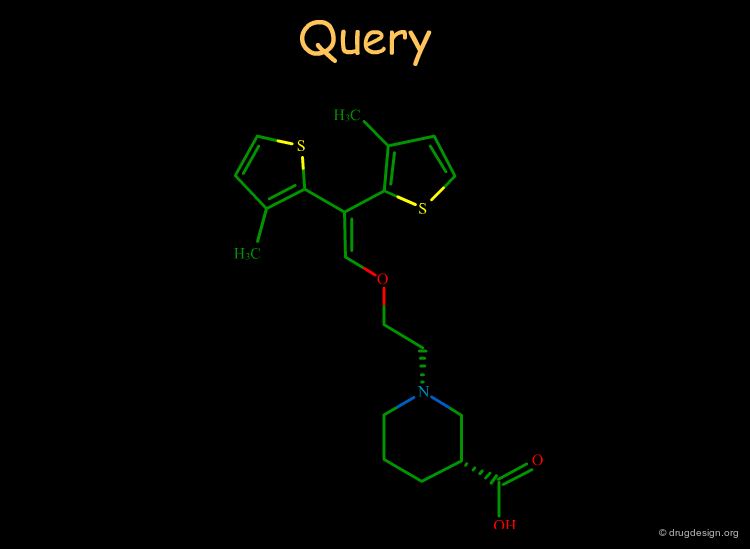
FlexS predicts ligand superpositions. For a given pair of ligands, it predicts the conformation and orientation of one of the ligands relative to the other one. Full torsional flexibility of each molecule is considered. For virtual screening one uses a reference structure as a negative fingerprint of the active site. FlexS estimates a score for the similarity between each database structure and the reference structure as part of the optimization of the alignment. In this way it provides a quantitative ranking of each database molecule.
ROCS¶
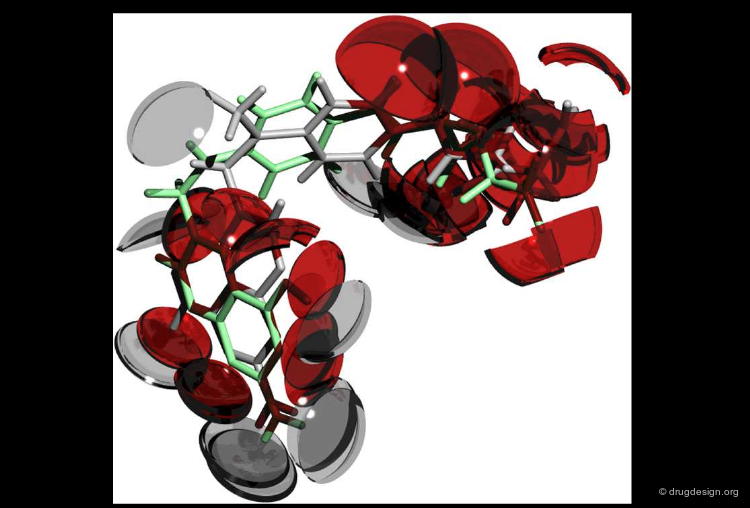
ROCS is a shape comparison program, based on the idea that molecules have similar shape if their volumes overlay well and that any volume mismatch is a measure of dissimilarity. It reports Tanimoto and Tversky measures of similarity between shapes. One can optionally specify a force field and its weighting to combine with shape matching. The query shape may be a molecule, a grid (e.g. electron density, active site, or arbitrary volume) or a composite of the two.
Copyright © 2024 drugdesign.org